








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 190-200.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0809
LIANG Ying-yi( ), ZHAO An-su, WANG Rui-xi, SHEN Tian-hong, MA Xin-rong, WANG Min, LUO Jian-mei(
), ZHAO An-su, WANG Rui-xi, SHEN Tian-hong, MA Xin-rong, WANG Min, LUO Jian-mei( )
)
Received:2025-07-26
Online:2025-11-26
Published:2025-12-09
Contact:
LUO Jian-mei
E-mail:908289174@qq.com;luojianmei@tust.edu.cn
LIANG Ying-yi, ZHAO An-su, WANG Rui-xi, SHEN Tian-hong, MA Xin-rong, WANG Min, LUO Jian-mei. Mining and Analysis of Proteins Associated with Neoxanthin Biosynthesis in Chlamydomonas reinhardtii[J]. Biotechnology Bulletin, 2025, 41(11): 190-200.
名称 Name | 引物序列 Primer sequence (5′-3′) | 用途 Application |
|---|---|---|
| aR-nxs1-F | 干扰藻株构建引物 | |
| aR-nxs1-R | 干扰藻株构建引物 | |
| aR-nxs2-F | 干扰藻株构建引物 | |
| aR-nxs2-R | 干扰藻株构建引物 | |
| aR-nxs3-F | 干扰藻株构建引物 | |
| aR-nxs3-R | 干扰藻株构建引物 | |
| AmiRNAprecF | GGTGTTGGGTCGGTGTTTTTG | 菌落PCR引物 |
| SpacerR | TAGCGCTGATCACCACCACCC | 菌落PCR引物 |
| Luciferase-F | CCATATGGTCAACGGCGTGAAGGTG | 干扰藻株初筛引物 |
| Luciferase-R | GCCGCCGGCGCCCTTGATCTTGTCC | 干扰藻株初筛引物 |
| nxs1-qF | TCATTCTCACCGTGGTAGCTGG | RT-qPCR引物 |
| nxs1-qR | AAGAACTGCGGGTTGAAGC | RT-qPCR引物 |
| nxs2-qF | GCTCACCCTGGATGTAGTCAT | RT-qPCR引物 |
| nxs2-qR | CGGTGGGTCCGAAGAAGAAG | RT-qPCR引物 |
| nxs3-qF | CGGACTTCAGCGAGATGTTC | RT-qPCR引物 |
| nxs3-qR | CAACCATCATGCACAGGAACAG | RT-qPCR引物 |
| α-tubulin-qF | CTCGCTTCGCTTTGACGGTG | 内参 |
| α-tubulin-qR | CGTGGTACGCCTTCTCGGC | 内参 |
Table 1 Primers used in this experiment
名称 Name | 引物序列 Primer sequence (5′-3′) | 用途 Application |
|---|---|---|
| aR-nxs1-F | 干扰藻株构建引物 | |
| aR-nxs1-R | 干扰藻株构建引物 | |
| aR-nxs2-F | 干扰藻株构建引物 | |
| aR-nxs2-R | 干扰藻株构建引物 | |
| aR-nxs3-F | 干扰藻株构建引物 | |
| aR-nxs3-R | 干扰藻株构建引物 | |
| AmiRNAprecF | GGTGTTGGGTCGGTGTTTTTG | 菌落PCR引物 |
| SpacerR | TAGCGCTGATCACCACCACCC | 菌落PCR引物 |
| Luciferase-F | CCATATGGTCAACGGCGTGAAGGTG | 干扰藻株初筛引物 |
| Luciferase-R | GCCGCCGGCGCCCTTGATCTTGTCC | 干扰藻株初筛引物 |
| nxs1-qF | TCATTCTCACCGTGGTAGCTGG | RT-qPCR引物 |
| nxs1-qR | AAGAACTGCGGGTTGAAGC | RT-qPCR引物 |
| nxs2-qF | GCTCACCCTGGATGTAGTCAT | RT-qPCR引物 |
| nxs2-qR | CGGTGGGTCCGAAGAAGAAG | RT-qPCR引物 |
| nxs3-qF | CGGACTTCAGCGAGATGTTC | RT-qPCR引物 |
| nxs3-qR | CAACCATCATGCACAGGAACAG | RT-qPCR引物 |
| α-tubulin-qF | CTCGCTTCGCTTTGACGGTG | 内参 |
| α-tubulin-qR | CGTGGTACGCCTTCTCGGC | 内参 |
| 时间Time (min) | 流速 Flow rate (mL/min) | A(%) | B(%) | C(%) |
|---|---|---|---|---|
| 0 | 1.0 | 100 | 0 | 0 |
| 4 | 1.0 | 0 | 100 | 0 |
| 18 | 1.0 | 0 | 20 | 80 |
| 21 | 1.0 | 0 | 100 | 0 |
| 29 | 1.0 | 100 | 0 | 0 |
Table 2 Liquid phase gradient elution conditions for neoxanthin
| 时间Time (min) | 流速 Flow rate (mL/min) | A(%) | B(%) | C(%) |
|---|---|---|---|---|
| 0 | 1.0 | 100 | 0 | 0 |
| 4 | 1.0 | 0 | 100 | 0 |
| 18 | 1.0 | 0 | 20 | 80 |
| 21 | 1.0 | 0 | 100 | 0 |
| 29 | 1.0 | 100 | 0 | 0 |
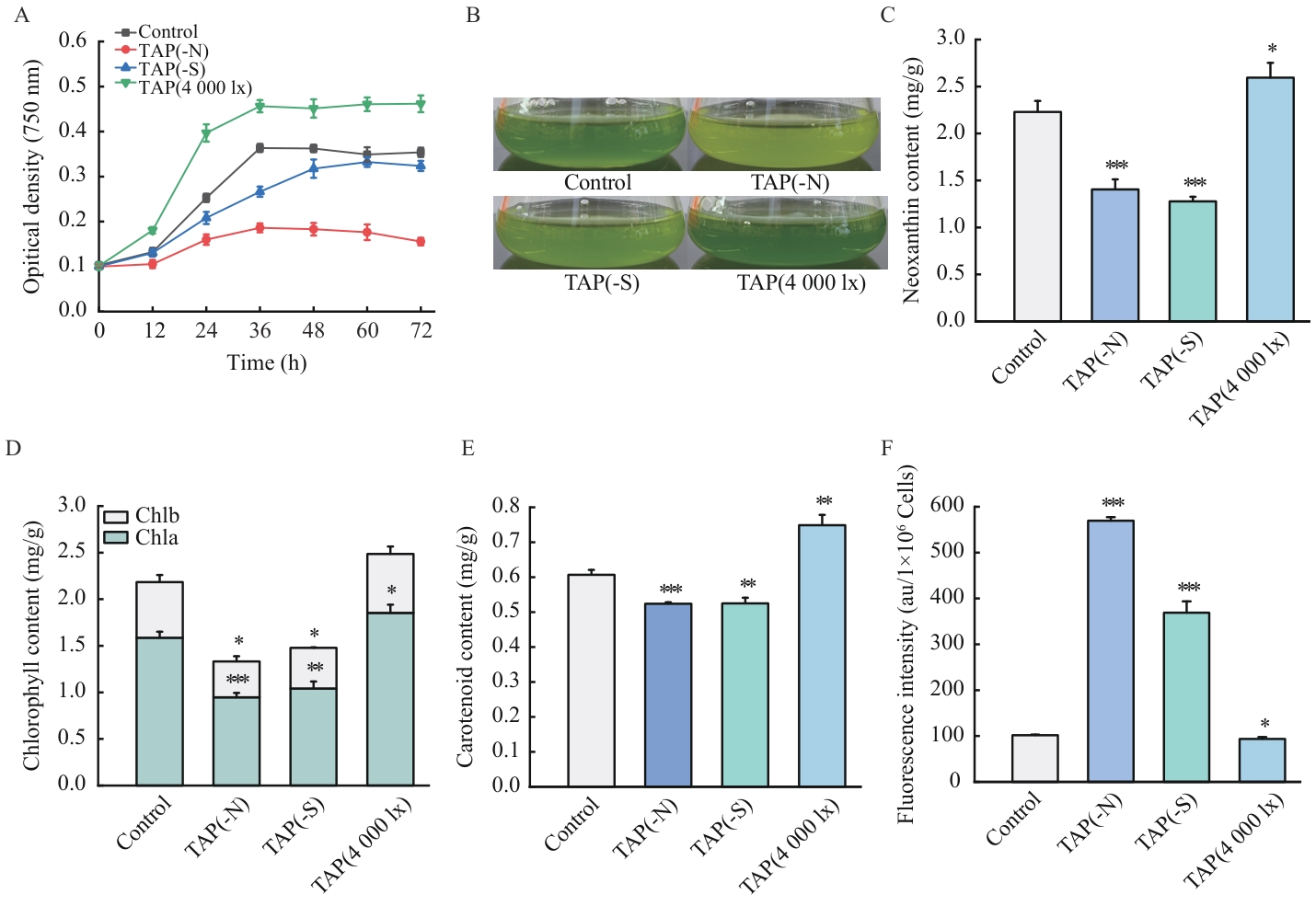
Fig. 1 Effects of culture conditions on the growth and pigment biosynthesis in C. reinhardtiiA: Growth curves. B: Culture status at mid-logarithmic phase. C: Neoxanthin content. D: Chlorophyll content. E: Carotenoid content. F: Lipid content. The values in the bar charts are of the mean±standard error (n=3); *, **, *** indicate P<0.05, P<0.01, P<0.001, respectively, the same below
基因 Gene | 染色体 Chromosome | 基因长度 Gene length (bp) | 染色体初始位置 Start locus on chromosome | 染色体结束位置 End locus on chromosome |
|---|---|---|---|---|
| nxs1 | 3 | 2 172 | 7289912 | 7292083 |
| nxs2 | 6 | 2 251 | 926797 | 929047 |
| nxs3 | 12 | 1 942 | 9172926 | 9174867 |
Table 3 Chromosomal locations of three neoxanthin synthase candidate genes in C. reinhardtii
基因 Gene | 染色体 Chromosome | 基因长度 Gene length (bp) | 染色体初始位置 Start locus on chromosome | 染色体结束位置 End locus on chromosome |
|---|---|---|---|---|
| nxs1 | 3 | 2 172 | 7289912 | 7292083 |
| nxs2 | 6 | 2 251 | 926797 | 929047 |
| nxs3 | 12 | 1 942 | 9172926 | 9174867 |
基因 Gene | 氨基酸长度 Amino acid length (aa) | 与文献鉴定的新黄素合成酶氨基酸序列一致性 Amino acid sequence identity to the identified neoxanthin synthase in literature (%) | DUF4281功能域位置 DUF4281 domain position | 与文献鉴定的新黄素合成酶的DUF4281的氨基酸序列一致性DUF4281 amino acid sequence identity to identified neoxanthin synthase in literature (%) | ||
|---|---|---|---|---|---|---|
拟南芥 A. thalian | 羽衣甘蓝 Chinese kale | 拟南芥 A. thalian | 羽衣甘蓝 Chinese kale | |||
| nxs1 | 258 | 49.1 | 48.9 | 91-222 | 55.3 | 54.9 |
| nxs2 | 226 | 46.8 | 46.6 | 75-214 | 46.2 | 46.2 |
| nxs3 | 105 | 21.5 | 21.8 | 3-72 | 31.1 | 31.1 |
Table 4 Analysis of amino acid sequences and functional domains between three candidate proteins in C. reinhardtii and A. thalian neoxanthin synthase
基因 Gene | 氨基酸长度 Amino acid length (aa) | 与文献鉴定的新黄素合成酶氨基酸序列一致性 Amino acid sequence identity to the identified neoxanthin synthase in literature (%) | DUF4281功能域位置 DUF4281 domain position | 与文献鉴定的新黄素合成酶的DUF4281的氨基酸序列一致性DUF4281 amino acid sequence identity to identified neoxanthin synthase in literature (%) | ||
|---|---|---|---|---|---|---|
拟南芥 A. thalian | 羽衣甘蓝 Chinese kale | 拟南芥 A. thalian | 羽衣甘蓝 Chinese kale | |||
| nxs1 | 258 | 49.1 | 48.9 | 91-222 | 55.3 | 54.9 |
| nxs2 | 226 | 46.8 | 46.6 | 75-214 | 46.2 | 46.2 |
| nxs3 | 105 | 21.5 | 21.8 | 3-72 | 31.1 | 31.1 |
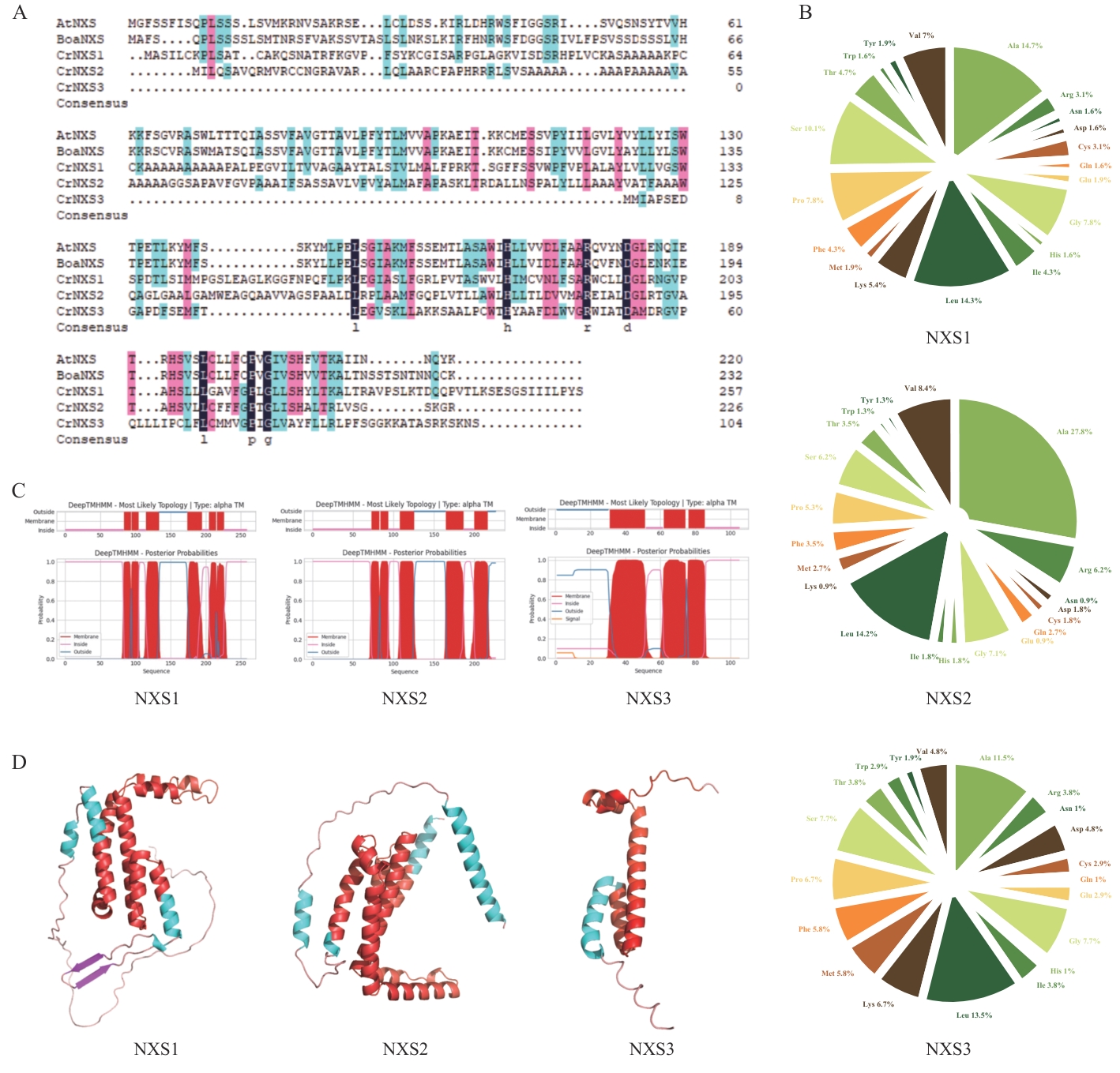
Fig. 2 Bioinformatics analysis of proteins associated with neoxanthin synthesisA: Amino acid sequence alignment of three candidate proteins in C. reinhardtii with the identified neoxanthin synthase in literature;AtNXS (neoxanthin synthase in A. thaliana), BoaNXS (neoxanthin synthase in Brassica oleracea var. acephala), CrNXS1-3 (neoxanthin synthase in C. reinhardtii). The conserved DUF4281 domain is boxed in red. B: Amino acid composition. C: Prediction analysis of transmembrane domain. D: The predicted tertiary structure is depicted as α-helices (green), β-sheets (pink), and the DUF4281 domain (red)
基因 Gene | 分子式 Molecular formula | 等电点 Isoelectric point (pI) | 不稳定指数 Instability index | 疏水性Hydropathicity (GRAVY) | 脂肪族指数 Aliphatic index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| nxs1 | C1228H1968N316O328S13 | 9.61 | 37.71 | 0.59 | 107.52 | Chloroplast |
| nxs2 | C1024H1659N289O272S10 | 10.77 | 42.27 | 0.863 | 114.38 | Plasma membrane |
| nxs3 | C520H816N130O137S9 | 8.91 | 46.29 | 0.357 | 92.98 | Chloroplast |
Table 5 Physicochemical properties and subcellular localization prediction of three candidate neoxanthin synthase proteins in C. reinhardtii
基因 Gene | 分子式 Molecular formula | 等电点 Isoelectric point (pI) | 不稳定指数 Instability index | 疏水性Hydropathicity (GRAVY) | 脂肪族指数 Aliphatic index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| nxs1 | C1228H1968N316O328S13 | 9.61 | 37.71 | 0.59 | 107.52 | Chloroplast |
| nxs2 | C1024H1659N289O272S10 | 10.77 | 42.27 | 0.863 | 114.38 | Plasma membrane |
| nxs3 | C520H816N130O137S9 | 8.91 | 46.29 | 0.357 | 92.98 | Chloroplast |
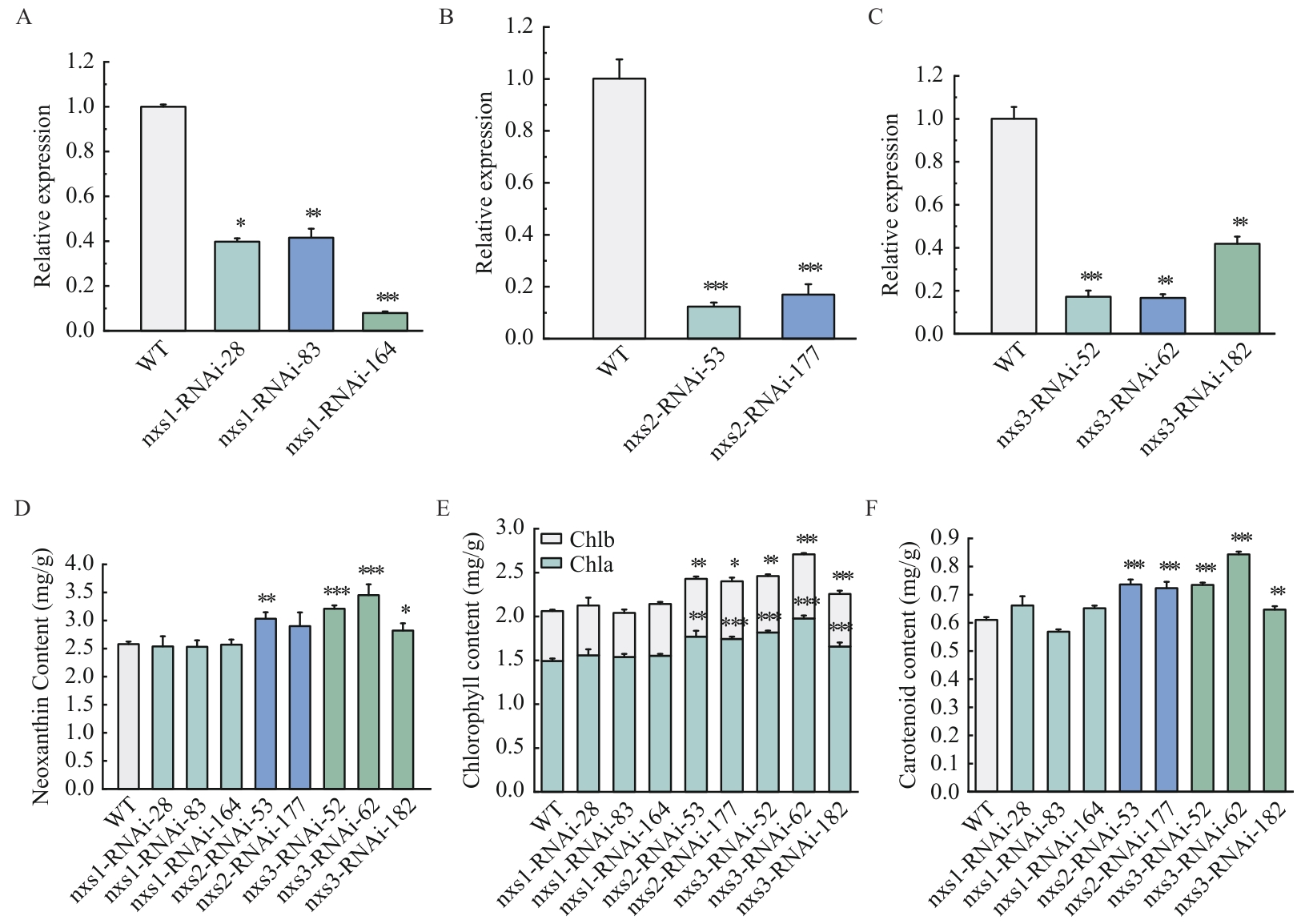
Fig. 3 Transcriptional level and pigment content of the RNAi strainsA: nxs1. B: nxs2. C: nxs3. D: neoxanthin content. E: Chlorophyll content. F: Carotenoid content
| [1] | Gauthier MR, Senhorinho GNA, Scott JA. Microalgae under environmental stress as a source of antioxidants [J]. Algal Res, 2020, 52: 102104. |
| [2] | Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants [J]. Trends Plant Sci, 2002, 7(1): 41-48. |
| [3] | Goldsmith TH, Krinsky NI. The epoxide nature of the carotenoid, neoxanthin [J]. Nature, 1960, 188: 491-493. |
| [4] | Cholnoky L, Györgyfy K, Rónai A, et al. Carotenoids and related compounds. Part XXI. Structure of neoxanthin (foliaxanthin) [J]. J Chem Soc C, 1969(9): 1256-1263. |
| [5] | Märki-Fischer E, Eugster CH. Neoflor und 6-epineoflor aus blüten von Trollius europaeus; hochfeld-1H-NMR-spektren von neoxanthin und (9'Z)-neoxanthin [J]. Helv Chim Acta, 1990, 73(6): 1637-1643. |
| [6] | Wang K, Tu WF, Liu C, et al. 9-Cis-neoxanthin in light harvesting complexes of photosystem II regulates the binding of violaxanthin and xanthophyll cycle [J]. Plant Physiol, 2017, 174(1): 86-96. |
| [7] | Giossi C, Cartaxana P, Cruz S. Photoprotective role of neoxanthin in plants and algae [J]. Molecules, 2020, 25(20): 4617. |
| [8] | Terasaki M. Potential ability of xanthophylls to prevent obesity-associated cancer [J]. World J Pharmacol, 2014, 3(4): 140. |
| [9] | Udayawara Rudresh D, Maradagi T, Stephen NM, et al. Neoxanthin prevents H2O2-induced cytotoxicity in HepG2 cells by activating endogenous antioxidant signals and suppressing apoptosis signals [J]. Mol Biol Rep, 2021, 48(10): 6923-6934. |
| [10] | Kotake-Nara E, Miyashita K, Nagao A, et al. Carotenoids affect proliferation of human prostate cancer cells [J]. J Nutr, 2001, 131(12): 3303-3306. |
| [11] | Kotake-Nara E, Asai A, Nagao A. Neoxanthin and fucoxanthin induce apoptosis in PC-3 human prostate cancer cells [J]. Cancer Lett, 2005, 220(1): 75-84. |
| [12] | Asai A, Terasaki M, Nagao A. An epoxide-furanoid rearrangement of spinach neoxanthin occurs in the gastrointestinal tract of mice and in vitro: formation and cytostatic activity of neochrome stereoisomers [J]. J Nutr, 2004, 134(9): 2237-2243. |
| [13] | Terasaki M, Asai A, Zhang H, et al. A highly polar xanthophyll of 9'-Cis-neoxanthin induces apoptosis in HCT116 human colon cancer cells through mitochondrial dysfunction [J]. Mol Cell Biochem, 2007, 300(1): 227-237. |
| [14] | Sekiya M, Suzuki S, Ushida Y, et al. Neoxanthin in young vegetable leaves prevents fat accumulation in differentiated adipocytes [J]. Biosci Biotechnol Biochem, 2021, 85(10): 2145-2152. |
| [15] | Phadwal K, Singh PK. Effect of nutrient depletion on β-carotene and glycerol accumulation in two strains of Dunaliella sp [J]. Bioresour Technol, 2003, 90(1): 55-58. |
| [16] | 黄昆美. 利用基因工程提高莱茵衣藻细胞中类胡萝卜素含量的研究[D]. 青岛: 青岛科技大学, 2022. |
| Huang KM. Study on improving the content of carotenoids in Chlamydomonas Reinhardtii cells by genetic engineering [D]. Qingdao: Qingdao University of Science & Technology, 2022. | |
| [17] | Becker C, Urlić B, Jukić Špika M, et al. Nitrogen limited red and green leaf lettuce accumulate flavonoid glycosides, caffeic acid derivatives, and sucrose while losing chlorophylls, Β-carotene and xanthophylls [J]. PLoS One, 2015, 10(11): e0142867. |
| [18] | North HM, De Almeida A, Boutin JP, et al. The Arabidopsis ABA-deficient mutant Aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers [J]. Plant J, 2007, 50(5): 810-824. |
| [19] | Jian Y, Zhang CL, Wang YT, et al. Characterization of the role of the neoxanthin synthase gene BoaNXS in carotenoid biosynthesis in Chinese kale [J]. Genes, 2021, 12(8): 1122. |
| [20] | Harris EH. CHLAMYDOMONAS as a model organism [J]. Annu Rev Plant Physiol Plant Mol Biol, 2001, 52: 363-406. |
| [21] | Yang L, Chen J, Qin S, et al. Growth and lipid accumulation by different nutrients in the microalga Chlamydomonas reinhardtii [J]. Biotechnol Biofuels, 2018, 11(1): 40. |
| [22] | Sakarika M, Kornaros M. Kinetics of growth and lipids accumulation in Chlorella vulgaris during batch heterotrophic cultivation: Effect of different nutrient limitation strategies [J]. Bioresour Technol, 2017, 243: 356-365. |
| [23] | Lv Y, Han F, Liu MX, et al. Characteristics of N 6-methyladenosine modification during sexual reproduction of Chlamydomonas reinhardtii [J]. Genom Proteom Bioinform, 2023, 21(4): 756-768. |
| [24] | Fabrowska J, Ibañez E, Łęska B, et al. Supercritical fluid extraction as a tool to valorize underexploited freshwater green algae [J]. Algal Res, 2016, 19: 237-245. |
| [25] | Akella S, Ma XR, Bacova R, et al. Co-targeting strategy for precise, scarless gene editing with CRISPR/Cas9 and donor ssODNs in Chlamydomonas [J]. Plant Physiol, 2021, 187(4): 2637-2655. |
| [26] | Nama S, Madireddi SK, Yadav RM, et al. Non-photochemical quenching-dependent acclimation and thylakoid organization of Chlamydomonas reinhardtii to high light stress [J]. Photosynth Res, 2019, 139(1): 387-400. |
| [27] | Wright SW, Jeffrey SW, Mantoura RFC, et al. Improved HPLC method for the analysis of chlorophylls and carotenoids from marine phytoplankton [J]. Mar Ecol Prog Ser, 1991, 77(2-3): 183-196. |
| [28] | Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents [J]. Biochem Soc Trans, 1983, 11(5): 591-592. |
| [29] | Zheng SY, Zou SY, Feng T, et al. Low temperature combined with high inoculum density improves alpha-linolenic acid production and biochemical characteristics of Chlamydomonas reinhardtii [J]. Bioresour Technol, 2022, 348: 126746. |
| [30] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [31] | Shang QY, Yan YX, Zhang YN, et al. Advanced microalgal cultivation in biomethanation digestate to maximize resource utilization of food waste leachate [J]. J Environ Manag, 2025, 391: 126379. |
| [32] | Hess SK, Lepetit B, Kroth PG, et al. Production of chemicals from microalgae lipids-status and perspectives [J]. Eur J Lipid Sci Technol, 2018, 120(1): 1700152. |
| [33] | Neuman H, Galpaz N, Cunningham FX Jr, et al. The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis [J]. Plant J, 2014, 78(1): 80-93. |
| [34] | Perreau F, Frey A, Effroy-Cuzzi D, et al. Abscisic acid-deficient4 has an essential function in both Cis-Violaxanthin and Cis-Neoxanthin synthesis [J]. Plant Physiol, 2020, 184(3): 1303-1316. |
| [1] | LI Ya, JIANG Lin, XU Chuang, WANG Su-hui, MA Zhao, WANG Liang. Research Progress in Molecular Defense Mechanisms of Chlamydomonas reinhardtii in Response to Heavy Metal Stress [J]. Biotechnology Bulletin, 2025, 41(8): 53-64. |
| [2] | LI Wang-ning, ZHANG Hao-jie, LI Ya-nan, LIANG Meng-jing, JI Chun-li, Zhang Chun-hui, LI Run-zhi, CUI Yu-lin, QIN Song, CUI Hong-li. Phenotypic Characterization of Blue Photoreceptor Plant Type Cryptochrome CRY Mutant in Chlamydomonas reinhardtii [J]. Biotechnology Bulletin, 2023, 39(2): 243-253. |
| [3] | LIU Xue-dan, YANG Meng, ZHANG Jing, ZHAO Dong-xu. Effects of Glucose-xylose Co-utilization on the Synthesis of D-1,2,4-Butanetriol by Recombinant Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(9): 171-179. |
| [4] | ZHANG Mei-jun, WU Qing, YIN Cui, WANG Ni, MA Xiao-qing, MA Xiao-xia, CAO Yun-e. Screening and Identification of an Antagonistic Strain Against Fusarium oxyporum f. sp. cucumerinum and Optimization of Culture Conditions [J]. Biotechnology Bulletin, 2020, 36(9): 125-136. |
| [5] | Chen Yabing, Lan Daoliang, Lin Baoshan, Huang Cai, Huang Yong, Li Jian. Relative Quantification of mRNA Transcription of TLR3 and TLR5 in Different Tissues of Yak [J]. Biotechnology Bulletin, 2014, 30(8): 89-93. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||