








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 124-131.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0069
Previous Articles Next Articles
TANG Yue-hui( ), ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen
), ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen
Received:2022-01-13
Online:2022-10-26
Published:2022-11-11
Contact:
TANG Yue-hui
E-mail:yhtang2005@163.com
TANG Yue-hui, ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen. Identification and Gene Mapping of a Seedling Lethal Mutant in Rice[J]. Biotechnology Bulletin, 2022, 38(10): 124-131.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Purpose |
|---|---|---|---|
| GUSF | GATGTTGGCGACCTCGTATT | 354 | 探针制备Probe preparation |
| GUSR | ACAGCGTCTCCGACCTGAT | ||
| eRF1-1F | GCTACTCCTACAAAACACCAGATG | 368 | |
| eRF1-1R | GCATACCAAGAAGGCAAGAAC | ||
| RAD1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA | Tail-PCR | |
| RAD2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT | ||
| RAD3 | ACGATGGACTCCAGAGCGGCCGCHNVNNNCCAC | ||
| RAD4 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA | ||
| RAD5 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT | ||
| AC | ACGATGGACTCCAGAG | ||
| RB1 | GTCGTCGGTGAACAGGTATGGAAT | ||
| RB2 | ACGATGGACTCCAGTCCGGCCTGGCGGTAACAAG- AAAGGGATCTTCACT | ||
| RB3 | AAACCGCAGCAGGGAGGCAAAC | ||
| OseRF1-1F | CGAGCACGACACGAGCTTT | 179 | 定量表达分析Quantitative expression analysis |
| OseRF1-1R | GTGCAGCATCAAGTCCTTTAAT | ||
| OseRF1-2F | AATCGTTCGTCCTGCTGCTT | 151 | |
| OseRF1-2R | TTGCCTCTAGCAGACTCCAGTG | ||
| OseRF1-3F | ACGAACATCACCGCAACCT | 178 | |
| OseRF1-3R | CGAAATCATGCTCGTACCGTT | ||
| OseRF1-4F | AAGTTCAGAGGCGAGGGTTT | 176 | |
| OseRF1-4R | CTGACTCCAGAGCCTTGATTAG | ||
| RUB1F | AGGGTTCACAAGTCTGCCTATT | 165 | |
| RUB1R | TTCCATGCTGCTCTACCACAG | ||
| OseRF1-1F OseRF1-1R | CGAGCACGACACGAGCTTT AGATCCAACGTCACAGGGTACAT | 1 342 | 亚细胞定位载体构建引物 Subcellular localization vector construction primers |
Table 1 Primers used in the study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Purpose |
|---|---|---|---|
| GUSF | GATGTTGGCGACCTCGTATT | 354 | 探针制备Probe preparation |
| GUSR | ACAGCGTCTCCGACCTGAT | ||
| eRF1-1F | GCTACTCCTACAAAACACCAGATG | 368 | |
| eRF1-1R | GCATACCAAGAAGGCAAGAAC | ||
| RAD1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA | Tail-PCR | |
| RAD2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT | ||
| RAD3 | ACGATGGACTCCAGAGCGGCCGCHNVNNNCCAC | ||
| RAD4 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA | ||
| RAD5 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT | ||
| AC | ACGATGGACTCCAGAG | ||
| RB1 | GTCGTCGGTGAACAGGTATGGAAT | ||
| RB2 | ACGATGGACTCCAGTCCGGCCTGGCGGTAACAAG- AAAGGGATCTTCACT | ||
| RB3 | AAACCGCAGCAGGGAGGCAAAC | ||
| OseRF1-1F | CGAGCACGACACGAGCTTT | 179 | 定量表达分析Quantitative expression analysis |
| OseRF1-1R | GTGCAGCATCAAGTCCTTTAAT | ||
| OseRF1-2F | AATCGTTCGTCCTGCTGCTT | 151 | |
| OseRF1-2R | TTGCCTCTAGCAGACTCCAGTG | ||
| OseRF1-3F | ACGAACATCACCGCAACCT | 178 | |
| OseRF1-3R | CGAAATCATGCTCGTACCGTT | ||
| OseRF1-4F | AAGTTCAGAGGCGAGGGTTT | 176 | |
| OseRF1-4R | CTGACTCCAGAGCCTTGATTAG | ||
| RUB1F | AGGGTTCACAAGTCTGCCTATT | 165 | |
| RUB1R | TTCCATGCTGCTCTACCACAG | ||
| OseRF1-1F OseRF1-1R | CGAGCACGACACGAGCTTT AGATCCAACGTCACAGGGTACAT | 1 342 | 亚细胞定位载体构建引物 Subcellular localization vector construction primers |

Fig. 1 Phenotype of wild type and ls mutant a:Phenotype of ls mutants at 3-5 leaf stage. b:Phenotype of stem base and sheath at seedling stage in wild type and ls mutant plants
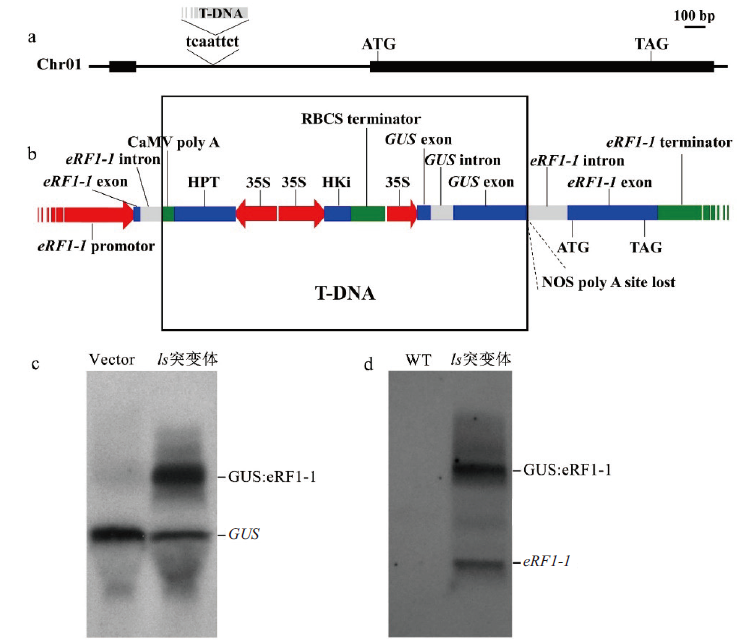
Fig. 5 T-DNA insertion site analysis of ls mutant a:T-DNA insertion site;b:T-DNA insert transcript analysis;c:GUS gene sequence was used as probe. Northern hybridization;d:eRF1-1 gene sequence was used as probe. Northern hybridization

Fig. 6 Analysis of amino acid sequences of eRF1 and its homologous proteins OseRF1-1:Oryza sativa Japonica,Os01g0939500;OseRF1-2:O. sativa,XP_015630898;OseRF1-3:O. sativa,XP_025881251;OseRF1-4:O. sativa,XP_015647295;AteRF1-1:Arabidopsis thaliana,AT5G47880;AteRF1-2:A. thaliana,AT1G12920;AteRF1-3:A. thaliana,AT3G26618;TaeRF1:Triticum aestivum,KAF6986654;ZmeRF1:Zea mays L.,ACG27998;AleRF1-3:Arabidopsis lyrata subsp. Lyrate,XP_002875327;BneRF1-3:Rhinolophus sinicus,XP_019577167
| [1] |
Mangkalaphiban K, He F, Ganesan R, et al. Transcriptome-wide investigation of stop codon readthrough in Saccharomyces cerevisiae[J]. PLoS Genet, 2021, 17(4):e1009538.
doi: 10.1371/journal.pgen.1009538 URL |
| [2] |
Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes—The impact of RNA sequence[J]. RNA Biol, 2015, 12(9):950-958.
doi: 10.1080/15476286.2015.1068497 pmid: 26176195 |
| [3] |
Crawford DJ, Ito K, Nakamura Y, et al. Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3[J]. EMBO J, 1999, 18(3):727-732.
pmid: 9927432 |
| [4] |
Saito K, Ito K. Genetic analysis of L123 of the tRNA-mimicking eukaryote release factor eRF1, an amino acid residue critical for discrimination of stop codons[J]. Nucleic Acids Res, 2015, 43(9):4591-4601.
doi: 10.1093/nar/gkv376 pmid: 25897120 |
| [5] |
Bertram G, Bell HA, Ritchie DW, et al. Terminating eukaryote translation:domain 1 of release factor eRF1 functions in stop codon recognition[J]. RNA, 2000, 6(9):1236-1247.
pmid: 10999601 |
| [6] |
Beiβel C, Neumann B, Uhse S, et al. Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3[J]. Nucleic Acids Res, 2019, 47(9):4798-4813.
doi: 10.1093/nar/gkz177 pmid: 30873535 |
| [7] |
Frolova LY, Tsivkovskii RY, Sivolobova GF, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis[J]. RNA, 1999, 5(8):1014-1020.
pmid: 10445876 |
| [8] |
Kurilla A, Szőke A, Auber A, et al. Expression of the translation termination factor eRF1 is autoregulated by translational readthrough and 3’UTR intron-mediated NMD in Neurospora crassa[J]. FEBS Lett, 2020, 594(21):3504-3517.
doi: 10.1002/1873-3468.13918 URL |
| [9] |
Nyikó T, Auber A, Szabadkai L, et al. Expression of the eRF1 translation termination factor is controlled by an autoregulatory circuit involving readthrough and nonsense-mediated decay in plants[J]. Nucleic Acids Res, 2017, 45(7):4174-4188.
doi: 10.1093/nar/gkw1303 pmid: 28062855 |
| [10] |
Yang Q, Yu CH, Zhao FZ, et al. eRF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons[J]. Nucleic Acids Res, 2019, 47(17):9243-9258.
doi: 10.1093/nar/gkz710 URL |
| [11] |
An Y, Lou YF, Xu YW. Overexpression, crystallization and preliminary X-ray crystallographic analysis of release factor eRF1-1 from Arabidopsis thaliana[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2013, 69(Pt 11):1295-1298.
doi: 10.1107/S1744309113027784 URL |
| [12] | Petsch KA, Mylne J, Botella JR. Cosuppression of eukaryotic release factor 1-1 in Arabidopsis affects cell elongation and radial cell division[J]. Plant Physiol, 2005, 139(1):115-126. |
| [13] |
Zhou XJ, Cooke P, Li L. Eukaryotic release factor 1-2 affects Arabidopsis responses to glucose and phytohormones during germination and early seedling development[J]. J Exp Bot, 2010, 61(2):357-367.
doi: 10.1093/jxb/erp308 URL |
| [14] |
Zhao B, Tang YY, Zhang BC, et al. The temperature-dependent retention of introns in GPI8 transcripts contributes to a drooping and fragile shoot phenotype in rice[J]. Int J Mol Sci, 2019, 21(1):299.
doi: 10.3390/ijms21010299 URL |
| [15] |
Codner GF, Erbs V, Loeffler J, et al. Universal Southern blot protocol with cold or radioactive probes for the validation of alleles obtained by homologous recombination[J]. Methods, 2021, 191:59-67.
doi: 10.1016/j.ymeth.2020.06.011 URL |
| [16] |
Yang C, Ma YM, He Y, et al. OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling[J]. Plant J, 2018, 93(3):489-501.
doi: 10.1111/tpj.13793 URL |
| [17] |
Zia MF, Flynt AS. Detection and verification of mammalian mirtrons by northern blotting[J]. Methods Mol Biol, 2018, 1823:209-219.
doi: 10.1007/978-1-4939-8624-8_16 pmid: 29959684 |
| [18] |
Tang YH, Li H, Guan YX, et al. Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5[J]. Front Genet, 2020, 11:670.
doi: 10.3389/fgene.2020.00670 URL |
| [19] |
Hellen CUT. Translation termination and ribosome recycling in eukaryotes[J]. Cold Spring Harb Perspect Biol, 2018, 10(10):a032656.
doi: 10.1101/cshperspect.a032656 URL |
| [20] |
Song H, Mugnier P, Das AK, et al. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis[J]. Cell, 2000, 100(3):311-321.
pmid: 10676813 |
| [21] |
Baradaran-Heravi A, Balgi AD, Hosseini-Farahabadi S, et al. Effect of small molecule eRF3 degraders on premature termination codon readthrough[J]. Nucleic Acids Res, 2021, 49(7):3692-3708.
doi: 10.1093/nar/gkab194 pmid: 33764477 |
| [22] |
Castellanos M, Mothi N, Muñoz V. Eukaryotic transcription factors can track and control their target genes using DNA antennas[J]. Nat Commun, 2020, 11(1):540.
doi: 10.1038/s41467-019-14217-8 pmid: 31992709 |
| [23] | 熊伟, 朱成新, 王玉婷, 等. CRISPR-Cas9介导靶向突变拟南芥ERF1-1基因[J]. 深圳大学学报:理工版, 2021, 38(5):504-509. |
| Xiong W, Zhu CX, Wang YT, et al. Targeted mutation of ERF1-1 gene in Arabidopsis using CRISPR-Cas9 gene editing system[J]. J Shenzhen Univ Sci Eng, 2021, 38(5):504-509. | |
| [24] |
Chen WC, Wang Q, Cao TJ, et al. UBC19 is a new interacting protein of ORANGE for its nuclear localization in Arabidopsis thaliana[J]. Plant Signal Behav, 2021, 16(11):1964847.
doi: 10.1080/15592324.2021.1964847 URL |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||