








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 80-89.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1616
Previous Articles Next Articles
XU Yuan-yuan1,2,3( ), ZHAO Guo-chun1,2,3, HAO Ying-ying1,2,3, WENG Xue-huang4, CHEN Zhong1,2,3,5(
), ZHAO Guo-chun1,2,3, HAO Ying-ying1,2,3, WENG Xue-huang4, CHEN Zhong1,2,3,5( ), JIA Li-ming1,2,3(
), JIA Li-ming1,2,3( )
)
Received:2021-12-30
Online:2022-10-26
Published:2022-11-11
Contact:
CHEN Zhong,JIA Li-ming
E-mail:yuanyuanxu_2016@163.com;zhongchen@bjfu.edu.cn;jlm@bjfu.edu.cn
XU Yuan-yuan, ZHAO Guo-chun, HAO Ying-ying, WENG Xue-huang, CHEN Zhong, JIA Li-ming. Reference Genes Selection and Validation for RT-qPCR in Sapindus mukorossi[J]. Biotechnology Bulletin, 2022, 38(10): 80-89.

Fig. 1 Sample materials of S. mukorossi R:root;ST:stem;L:leaf;B:bud;MF:male flower;FF:female flower;S1:pericarp at 15 d after flower pollination;S2:pericarp at 45 d after flower pollination;S3:pericarp at 75 d after flower pollination;S4:pericarp at 90 d after flower pollination;S5:pericarp at 105 d after flower pollination;S6:pericarp at 120 d after flower pollination;S7:pericarp at 135 d after flower pollination;S8:pericarp at 150 d after flower pollination
| 内参基因 Reference genes | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequences(5'- 3') |
|---|---|---|---|---|
| Sm18S | Samuk09G0136600 | 221 | AGATTGAAGGCGTTCTTTGGAT | TCAGTGTTTATGGACGGTGGAC |
| SmACT | Samuk13G0061200 | 129 | AGAAAGTTGGCCTCGCTGAA | CAGGAACCAGACCACCTGTC |
| SmEF-1α | Samuk14G0055700 | 101 | TCCAAGGCCAGGTACGATGA | AAACCAGAGATGGGGACGAAG |
| SmRPL1 | Samuk07G0011500 | 206 | CTGACCACCCCACCACTCTC | TCCTCCTCATCTGCCACCTC |
| SmRPS26 | Samuk08G0043900 | 230 | GCGAAGAAATGGAGGAAGGA | AGTGGATGGCACACGAAACA |
| SmTBCC | Samuk01G0035500 | 130 | AGCCTGACACCCTTTCTCTACC | CCAACCTCTTCTGCAGCCTT |
| SmUBC12 | Samuk11G0037500 | 102 | TTGTCTGGACCAACCAAAGGA | TGCTTCTTGCCAGGTGTTTTC |
| SmUBP | Samuk09G0089700 | 180 | TGCACAGTTGTTGCTCATGC | CTGCTTCATTTTCCCCGTGC |
Table 1 Primers of candidate reference genes
| 内参基因 Reference genes | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequences(5'- 3') |
|---|---|---|---|---|
| Sm18S | Samuk09G0136600 | 221 | AGATTGAAGGCGTTCTTTGGAT | TCAGTGTTTATGGACGGTGGAC |
| SmACT | Samuk13G0061200 | 129 | AGAAAGTTGGCCTCGCTGAA | CAGGAACCAGACCACCTGTC |
| SmEF-1α | Samuk14G0055700 | 101 | TCCAAGGCCAGGTACGATGA | AAACCAGAGATGGGGACGAAG |
| SmRPL1 | Samuk07G0011500 | 206 | CTGACCACCCCACCACTCTC | TCCTCCTCATCTGCCACCTC |
| SmRPS26 | Samuk08G0043900 | 230 | GCGAAGAAATGGAGGAAGGA | AGTGGATGGCACACGAAACA |
| SmTBCC | Samuk01G0035500 | 130 | AGCCTGACACCCTTTCTCTACC | CCAACCTCTTCTGCAGCCTT |
| SmUBC12 | Samuk11G0037500 | 102 | TTGTCTGGACCAACCAAAGGA | TGCTTCTTGCCAGGTGTTTTC |
| SmUBP | Samuk09G0089700 | 180 | TGCACAGTTGTTGCTCATGC | CTGCTTCATTTTCCCCGTGC |
| 内参基因 Reference gene | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequence(5'- 3') |
|---|---|---|---|---|
| SmAACT4 | Samuk13G0079600 | 132 | CCTGTTTTGAGGGCATTGATTG | ACCAGCATAGAACGCAGCCA |
| SmDXS4 | Samuk14G0066700 | 194 | AACAATGCTCAGAAGCCGAGA | GCGATCCATCCAGTAATCCAC |
| SmFPS | Samuk04G0156300 | 131 | GAGGCAGAAGTAGGAACTCACCA | CCATTGTAGCAGTCAAGTGGTAAG |
| SmbAS1 | Samuk02G0323400 | 113 | GAGTGGGATACTGGTTTCGCTAT | TCCTTGACCTGAGATGCTTTGA |
| SmCYP716A-5 | Samuk08G0024600 | 146 | GCTGTTTTCTGTGGCCCTTC | CGAGTCTTGGCAATTTTCCC |
| SmUGT73C-14 | Samuk12G0007500 | 120 | ACCTACAAGTGCCCAAACAGAT | ATTCCCAGTTCACCACATTCC |
| SmbHLH8 | Samuk10G0068800 | 107 | CAACCTTTGACTCCGACTCCA | CCTTCCCTTACCCTAACCTCC |
| SmERF25 | Samuk11G0050000 | 129 | CAACAGCCACACCAACAGCA | TCTTCTCGGGTCACGGATCTC |
Table 2 Primers of objective genes
| 内参基因 Reference gene | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequence(5'- 3') |
|---|---|---|---|---|
| SmAACT4 | Samuk13G0079600 | 132 | CCTGTTTTGAGGGCATTGATTG | ACCAGCATAGAACGCAGCCA |
| SmDXS4 | Samuk14G0066700 | 194 | AACAATGCTCAGAAGCCGAGA | GCGATCCATCCAGTAATCCAC |
| SmFPS | Samuk04G0156300 | 131 | GAGGCAGAAGTAGGAACTCACCA | CCATTGTAGCAGTCAAGTGGTAAG |
| SmbAS1 | Samuk02G0323400 | 113 | GAGTGGGATACTGGTTTCGCTAT | TCCTTGACCTGAGATGCTTTGA |
| SmCYP716A-5 | Samuk08G0024600 | 146 | GCTGTTTTCTGTGGCCCTTC | CGAGTCTTGGCAATTTTCCC |
| SmUGT73C-14 | Samuk12G0007500 | 120 | ACCTACAAGTGCCCAAACAGAT | ATTCCCAGTTCACCACATTCC |
| SmbHLH8 | Samuk10G0068800 | 107 | CAACCTTTGACTCCGACTCCA | CCTTCCCTTACCCTAACCTCC |
| SmERF25 | Samuk11G0050000 | 129 | CAACAGCCACACCAACAGCA | TCTTCTCGGGTCACGGATCTC |
| 内参基因 Reference gene | 扩增效率 Amplification efficiency /% | 相关系数R2 Correlation coefficient R2 | 斜率K Slope K |
|---|---|---|---|
| Sm18S | 97.872 | 0.998 | -3.374 |
| SmACT | 93.727 | 0.999 | -3.482 |
| SmEF-1α | 99.789 | 0.999 | -3.327 |
| SmRPL1 | 100.206 | 0.999 | -3.317 |
| SmRPS26 | 94.469 | 0.999 | -3.462 |
| SmTBCC | 97.196 | 0.997 | -3.391 |
| SmUBC12 | 96.881 | 0.999 | -3.399 |
| SmUBP | 98.032 | 0.999 | -3.370 |
Table 3 Amplification efficiency and standard curve para-meters of candidate reference genes
| 内参基因 Reference gene | 扩增效率 Amplification efficiency /% | 相关系数R2 Correlation coefficient R2 | 斜率K Slope K |
|---|---|---|---|
| Sm18S | 97.872 | 0.998 | -3.374 |
| SmACT | 93.727 | 0.999 | -3.482 |
| SmEF-1α | 99.789 | 0.999 | -3.327 |
| SmRPL1 | 100.206 | 0.999 | -3.317 |
| SmRPS26 | 94.469 | 0.999 | -3.462 |
| SmTBCC | 97.196 | 0.997 | -3.391 |
| SmUBC12 | 96.881 | 0.999 | -3.399 |
| SmUBP | 98.032 | 0.999 | -3.370 |

Fig. 2 Ct values of candidate reference genes The upper and lower parts of the box whiskers indicate the maximum and minimum values of the Ct value;the upper and lower parts of the box body indicate the upper and lower quartiles of the Ct value;lines across the boxes depict the medians;solid diamonds represent outliers and hollow squares represent averages
| 内参基因 Reference gene | geNorm | NormFinder | BestKeeper | Delta Ct | RefFinder综合排序 Comprehensive ranking | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M值 M value | 排序 Rank | SV值 SV value | 排序 Rank | SD值 SD value | 排序 Rank | CV 值 CV value/% | 排序 Rank | r | 排序 Rank | 平均标准偏差 STDEV | 排序 Rank | 基因稳定性Gene stability | 排序 Rank | ||||||
| Sm18S | 0.710 | 6 | 0.722 | 7 | 1.41 | 1 | 5.71 | 4 | 0.950 | 5 | 0.900 | 7 | 4.300 | 5 | |||||
| SmACT | 0.373 | 1 | 0.347 | 1 | 1.51 | 4 | 6.04 | 5 | 0.985 | 1 | 0.670 | 1 | 1.410 | 1 | |||||
| SmEF-1α | 0.799 | 7 | 0.943 | 8 | 1.54 | 6 | 7.19 | 7 | 0.922 | 6 | 1.070 | 8 | 7.740 | 8 | |||||
| SmRPL1 | 0.373 | 1 | 0.373 | 3 | 1.53 | 5 | 5.65 | 3 | 0.985 | 1 | 0.680 | 2 | 2.450 | 2 | |||||
| SmRPS26 | 0.656 | 5 | 0.666 | 6 | 1.68 | 7 | 7.65 | 8 | 0.982 | 2 | 0.860 | 6 | 6.450 | 7 | |||||
| SmTBCC | 0.615 | 4 | 0.501 | 5 | 1.45 | 3 | 5.18 | 1 | 0.973 | 4 | 0.770 | 5 | 4.400 | 6 | |||||
| SmUBC12 | 0.508 | 2 | 0.480 | 4 | 1.53 | 5 | 6.31 | 6 | 0.979 | 3 | 0.740 | 4 | 3.940 | 4 | |||||
| SmUBP | 0.547 | 3 | 0.362 | 2 | 1.43 | 2 | 5.59 | 2 | 0.985 | 1 | 0.700 | 3 | 2.630 | 3 | |||||
Table 4 GeNorm,NormFinder and BestKeeper analysis results and rankings
| 内参基因 Reference gene | geNorm | NormFinder | BestKeeper | Delta Ct | RefFinder综合排序 Comprehensive ranking | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M值 M value | 排序 Rank | SV值 SV value | 排序 Rank | SD值 SD value | 排序 Rank | CV 值 CV value/% | 排序 Rank | r | 排序 Rank | 平均标准偏差 STDEV | 排序 Rank | 基因稳定性Gene stability | 排序 Rank | ||||||
| Sm18S | 0.710 | 6 | 0.722 | 7 | 1.41 | 1 | 5.71 | 4 | 0.950 | 5 | 0.900 | 7 | 4.300 | 5 | |||||
| SmACT | 0.373 | 1 | 0.347 | 1 | 1.51 | 4 | 6.04 | 5 | 0.985 | 1 | 0.670 | 1 | 1.410 | 1 | |||||
| SmEF-1α | 0.799 | 7 | 0.943 | 8 | 1.54 | 6 | 7.19 | 7 | 0.922 | 6 | 1.070 | 8 | 7.740 | 8 | |||||
| SmRPL1 | 0.373 | 1 | 0.373 | 3 | 1.53 | 5 | 5.65 | 3 | 0.985 | 1 | 0.680 | 2 | 2.450 | 2 | |||||
| SmRPS26 | 0.656 | 5 | 0.666 | 6 | 1.68 | 7 | 7.65 | 8 | 0.982 | 2 | 0.860 | 6 | 6.450 | 7 | |||||
| SmTBCC | 0.615 | 4 | 0.501 | 5 | 1.45 | 3 | 5.18 | 1 | 0.973 | 4 | 0.770 | 5 | 4.400 | 6 | |||||
| SmUBC12 | 0.508 | 2 | 0.480 | 4 | 1.53 | 5 | 6.31 | 6 | 0.979 | 3 | 0.740 | 4 | 3.940 | 4 | |||||
| SmUBP | 0.547 | 3 | 0.362 | 2 | 1.43 | 2 | 5.59 | 2 | 0.985 | 1 | 0.700 | 3 | 2.630 | 3 | |||||
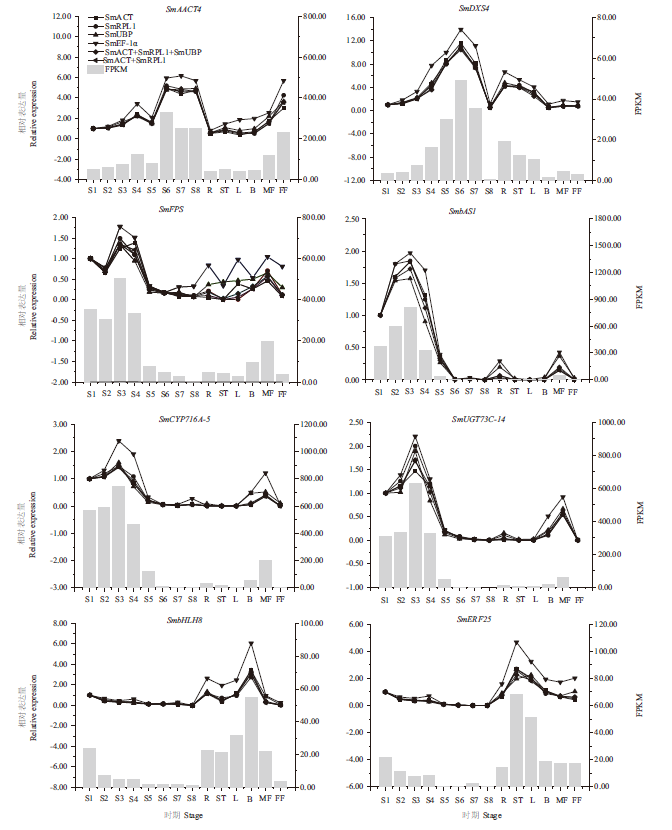
Fig. 4 RT-qPCR expression pattern of genes related to the triterpenoid saponin biosynthesis of S. mukorossi R,root;ST,stem;L,leaf;B,bud;MF,male flower;FF,female flower;S1,pericarp at 15 d after flower pollination;S2,pericarp at 45 d after flower pollination;S3,pericarp at 75 d after flower pollination;S4,pericarp at 90 d after flower pollination;S5,pericarp at 105 d after flower pollination;S6,pericarp at 120 d after flower pollination;S7,pericarp at 135 d after flower pollination;S8,pericarp at 150 d after flower pollination
| [1] |
Kou XY, Zhang L, Yang SZ, et al. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions[J]. Sci Hortic, 2017, 225:195-203.
doi: 10.1016/j.scienta.2017.07.004 URL |
| [2] |
Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines:minimum information for publication of quantitative real-time PCR experiments[J]. Clin Chem, 2009, 55(4):611-622.
doi: 10.1373/clinchem.2008.112797 URL |
| [3] |
Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR[J]. Nat Protoc, 2006, 1(3):1559-1582.
doi: 10.1038/nprot.2006.236 pmid: 17406449 |
| [4] |
Xu LF, Xu H, Cao YW, et al. Validation of reference genes for quantitative real-time PCR during bicolor tepal development in Asiatic hybrid lilies(Lilium spp. )[J]. Front Plant Sci, 2017, 8:669.
doi: 10.3389/fpls.2017.00669 URL |
| [5] |
Ferradás Y, Rey L, Martínez Ó, et al. Identification and validation of reference genes for accurate normalization of real-time quantitative PCR data in kiwifruit[J]. Plant Physiol Biochem, 2016, 102:27-36.
doi: 10.1016/j.plaphy.2016.02.011 URL |
| [6] |
Wu JY, Zhang HN, Liu LQ, et al. Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest longan fruits under different experimental conditions[J]. Front Plant Sci, 2016, 7:780.
doi: 10.3389/fpls.2016.00780 pmid: 27375640 |
| [7] |
袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2012, 47(4):427-436.
doi: 10.3724/SP.J.1259.2012.00427 |
| Yuan W, Wan HJ, Yang YJ. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants[J]. Chin Bull Bot, 2012, 47(4):427-436. | |
| [8] | 王丽平, 梁瑾, 谌琴琴, 等. 千里光RT-qPCR分析中内参基因的选择[J]. 中国中药杂志, 2019, 44(3):465-471. |
| Wang LP, Liang J, Shen QQ, et al. Identification of stable reference gene by RT-qPCR in Senecio scandens[J]. China J Chin Mater Med, 2019, 44(3):465-471. | |
| [9] | 徐碧霞, 郭巧生, 朱再标, 等. 老鸦瓣实时定量PCR内参基因的筛选和验证[J]. 中国中药杂志, 2021, 46(4):938-943. |
| Xu BX, Guo QS, Zhu ZB, et al. Selection and validation of reference genes for quantitative Real-time PCR analysis in Amana edulis[J]. China J Chin Mater Med, 2021, 46(4):938-943. | |
| [10] |
Li DD, Hu B, Wang Q, et al. Identification and evaluation of reference genes for accurate transcription normalization in safflower under different experimental conditions[J]. PLoS One, 2015, 10(10):e0140218.
doi: 10.1371/journal.pone.0140218 URL |
| [11] |
Nicot N, Hausman JF, Hoffmann L, et al. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress[J]. J Exp Bot, 2005, 56(421):2907-2914.
pmid: 16188960 |
| [12] | 贾黎明, 孙操稳. 生物柴油树种无患子研究进展[J]. 中国农业大学学报, 2012, 17(6):191-196. |
| Jia LM, Sun CW. Research progress of biodiesel tree Sapindus mukorossi[J]. J China Agric Univ, 2012, 17(6):191-196. | |
| [13] | 徐圆圆, 周思维, 陈仲, 等. 无患子不同器官中的总皂苷和总黄酮含量[J]. 南京林业大学学报:自然科学版, 2021, 45(4):83-89. |
| Xu YY, Zhou SW, Chen Z, et al. Contents of the total saponins and total flavonoids in different organs of Sapindus mukorossi[J]. J Nanjing For Univ Nat Sci Ed, 2021, 45(4):83-89. | |
| [14] | 刘济铭, 陈仲, 孙操稳, 等. 无患子属种质资源种实性状变异及综合评价[J]. 林业科学, 2019, 55(6):44-54. |
| Liu JM, Chen Z, Sun CW, et al. Variation in fruit and seed properties and comprehensive assessment of germplasm resources of the genus Sapindus[J]. Sci Silvae Sin, 2019, 55(6):44-54. | |
| [15] | 徐圆圆, 贾黎明, 陈仲, 等. 无患子三萜皂苷研究进展[J]. 化学通报, 2018, 81(12):1078-1088. |
| Xu YY, Jia LM, Chen Z, et al. Advances on triterpenoid saponin of Sapindus mukorossi[J]. Chemistry, 2018, 81(12):1078-1088. | |
| [16] |
Sun CW, Wang LC, Liu JM, et al. Genetic structure and biogeographic divergence among Sapindus species:an inter-simple sequence repeat-based study of germplasms in China[J]. Ind Crops Prod, 2018, 118:1-10.
doi: 10.1016/j.indcrop.2018.03.029 URL |
| [17] |
Zhao GC, Gao YH, Gao SL, et al. The phenological growth stages of Sapindus mukorossi according to BBCH scale[J]. Forests, 2019, 10(6):462.
doi: 10.3390/f10060462 URL |
| [18] |
Gao Y, Gao SL, Jia LM, et al. Canopy characteristics and light distribution in Sapindus mukorossi Gaertn. are influenced by crown architecture manipulation in the hilly terrain of Southeast China[J]. Sci Hortic, 2018, 240:11-22.
doi: 10.1016/j.scienta.2018.05.034 URL |
| [19] |
Hu QW, Chen YY, Jiao QY, et al. Triterpenoid saponins from the pulp of Sapindus mukorossi and their antifungal activities[J]. Phytochemistry, 2018, 147:1-8.
doi: 10.1016/j.phytochem.2017.12.004 URL |
| [20] | 陈莎莎, 陈丽萍, 林继辉. 无患子果皮皂苷提取工艺研究[J]. 云南民族大学学报:自然科学版, 2019, 28(6):558-562. |
| Chen SS, Chen LP, Lin JH. The extraction technology of saponins from the soapberry pericarp[J]. J Yunnan Minzu Univ Nat Sci Ed, 2019, 28(6):558-562. | |
| [21] |
Xu YY, Gao Y, Chen Z, et al. Metabolomics analysis of the soapberry(Sapindus mukorossi Gaertn.)pericarp during fruit development and ripening based on UHPLC-HRMS[J]. Sci Rep, 2021, 11(1):11657.
doi: 10.1038/s41598-021-91143-0 URL |
| [22] | Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biol, 2002, 3(7):RESEARCH0034. |
| [23] |
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data:a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Res, 2004, 64(15):5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 URL |
| [24] |
Pfaffl MW, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity:BestKeeper—Excel-based tool using pair-wise correlations[J]. Biotechnol Lett, 2004, 26(6):509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [25] |
Xie F, Xiao P, Chen D, et al. miRDeepFinder:a miRNA analysis tool for deep sequencing of plant small RNAs[J]. Plant Mol Biol, 2012. DOI: 10.1007/s11103-012-9885-2.
doi: 10.1007/s11103-012-9885-2 |
| [26] | 蒋婷婷, 高燕会, 童再康. 石蒜属植物实时荧光定量PCR内参基因的选择[J]. 园艺学报, 2015, 42(6):1129-1138. |
| Jiang TT, Gao YH, Tong ZK. Selection of reference genes for quantitative real-time PCR in Lycoris[J]. Acta Hortic Sin, 2015, 42(6):1129-1138. | |
| [27] |
Dudziak K, Sozoniuk M, Szczerba H, et al. Identification of stable reference genes for qPCR studies in common wheat(Triticum aestivum L.)seedlings under short-term drought stress[J]. Plant Methods, 2020, 16:58.
doi: 10.1186/s13007-020-00601-9 URL |
| [28] | 曹映辉, 郑燕, 张燕萍, 等. 建兰花香物质合成相关基因RT-qPCR内参基因筛选[J]. 分子植物育种, 2021, http://kns.cnki.net/kcms/detail/46.1068.S.20210510.1555.008.html. |
| Cao YH, Zheng Y, Zhang YP, et al. Selection of suitable RT-qPCR reference genes for floral scent biosynthesis in Cymbidium ensifolium[J]. Mol Plant Breed, 2021, http://kns.cnki.net/kcms/detail/46.1068.S.20210510.1555.008.html. | |
| [29] | 章颖佳, 程少禹, 王卓为, 等. 紫玉兰‘红元宝’花芽分化阶段基因定量分析的内参基因筛选[J]. 广西植物, 2022, 42(1):113-121. |
| Zhang YJ, Cheng SY, Wang ZW, et al. Selection of reference genes in Magnolia liliflora ‘Hongyuanbao’ during flower bud differentiation[J]. Guihaia, 2022, 42(1):113-121. | |
| [30] |
de Spiegelaere W, Dern-Wieloch J, Weigel R, et al. Reference gene validation for RT-qPCR, a note on different available software packages[J]. PLoS One, 2015, 10(3):e0122515.
doi: 10.1371/journal.pone.0122515 URL |
| [31] | 张玉芳, 赵丽娟, 曾幼玲. 基因表达研究中内参基因的选择与应用[J]. 植物生理学报, 2014, 50(8):1119-1125. |
| Zhang YF, Zhao LJ, Zeng YL. Selection and application of reference genes for gene expression studies[J]. Plant Physiol J, 2014, 50(8):1119-1125. | |
| [32] |
Hwang HS, Lee H, Choi YE. Transcriptomic analysis of Siberian ginseng(Eleutherococcus senticosus)to discover genes involved in saponin biosynthesis[J]. BMC Genomics, 2015, 16(1):180.
doi: 10.1186/s12864-015-1357-z URL |
| [33] |
Han JY, Chun JH, Oh SA, et al. Transcriptomic analysis of Kalopanax septemlobus and characterization of KsBAS, CYP716A94 and CYP72A397 genes involved in hederagenin saponin biosynthesis[J]. Plant Cell Physiol, 2018, 59(2):319-330.
doi: 10.1093/pcp/pcx188 URL |
| [34] |
Shan CM, Wang CK, Zhang SX, et al. Transcriptome analysis of Clinopodium gracile(Benth.)Matsum and identification of genes related to Triterpenoid Saponin biosynthesis[J]. BMC Genomics, 2020, 21(1):49.
doi: 10.1186/s12864-020-6454-y URL |
| [35] | Xiao Z, Sun XB, Liu XQ, et al. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. don[J]. Front Plant Sci, 2016, 7:1547. |
| 徐圆圆, 陈仲, 贾黎明, 等. 植物三萜皂苷生物合成途径及调控机制研究进展[J]. 中国科学:生命科学, 2021, 51(5):525-555. | |
|
Xu YY, Chen Z, Jia LM, et al. Advances in understanding of the biosynthetic pathway and regulatory mechanism of triterpenoid saponins in plants[J]. Sci Sin Vitae, 2021, 51(5):525-555.
doi: 10.1360/SSV-2020-0230 URL |
|
| [37] |
Zhao YJ, Li C. Biosynthesis of plant triterpenoid saponins in microbial cell factories[J]. J Agric Food Chem, 2018, 66(46):12155-12165.
doi: 10.1021/acs.jafc.8b04657 URL |
| [38] |
Augustin JM, Kuzina V, Andersen SB, et al. Molecular activities, biosynthesis and evolution of triterpenoid saponins[J]. Phytochemistry, 2011, 72(6):435-457.
doi: 10.1016/j.phytochem.2011.01.015 pmid: 21333312 |
| [39] |
Xu YY, Zhao GC, Ji XQ, et al. Metabolome and transcriptome analysis reveals the transcriptional regulatory mechanism of triterpenoid saponin biosynthesis in soapberry(Sapindus mukorossi Gaertn.)[J]. J Agric Food Chem, 2022, DOI: 10.1021/acs.jafc.2c01672.
doi: 10.1021/acs.jafc.2c01672 |
| [1] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [2] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [3] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [4] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [5] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [6] | SONG Hai-na, WU Xin-tong, YANG Lu-yu, GENG Xi-ning, ZHANG Hua-min, SONG Xiao-long. Selection and Validation of Reference Genes for RT-qPCR in Allium tuberosum Infected by Botrytis squamosa [J]. Biotechnology Bulletin, 2023, 39(3): 101-115. |
| [7] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [8] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [9] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [10] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [11] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [12] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [13] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [14] | YOU Chui-huai, XIE Jin-jin, ZHANG Ting, CUI Tian-zhen, SUN Xin-lu, ZANG Shou-jian, WU Yi-ning, SUN Meng-yao, QUE You-xiong, SU Ya-chun. Identification of the Lipoxygenase Gene GeLOX1 and Expression Analysis Under Low Temperature Stress in Gelsmium elegans [J]. Biotechnology Bulletin, 2023, 39(11): 318-327. |
| [15] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||