








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (8): 233-243.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1389
Previous Articles Next Articles
CHEN Ying( ), WANG Yi-lei, ZOU Peng-fei(
), WANG Yi-lei, ZOU Peng-fei( )
)
Received:2021-11-05
Online:2022-08-26
Published:2022-09-14
Contact:
ZOU Peng-fei
E-mail:chenying1445@163.com;pengfeizou@jmu.edu.cn
CHEN Ying, WANG Yi-lei, ZOU Peng-fei. Cloning and Expression Analysis of TRAF6 from Large Yellow Croaker Larimichthys crocea[J]. Biotechnology Bulletin, 2022, 38(8): 233-243.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 目的Application |
|---|---|---|
| Lc-TRAF6-F | ATGGCTTGCATTGACAGCAAT | Lc-TRAF6 ORF cloning |
| Lc-TRAF6-R | AGCCTCGGTTTCAAGGGAC | |
| pTurbo-TRAF6-F | CCGGAATTCTGATGGCTTGCATTGACAGCAAT | pTurbo-TRAF6-GFP |
| pTurbo-TRAF6-R | CGCGGATCCCGGTCCCTTGAAACCGAGGCT | |
| qTRAF6-F | GACGGACGGTTGGTAAAGCAG | RT-qPCR |
| qTRAF6-R | CAACTTGTAGCCTGGACGACCC | |
| qβ-actin-F | TTATGAAGGCTATGCCCTGCC | RT-qPCR |
| qβ-actin-R | TGAAGGAGTAGCCACGCTCTGT |
Table 1 Primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 目的Application |
|---|---|---|
| Lc-TRAF6-F | ATGGCTTGCATTGACAGCAAT | Lc-TRAF6 ORF cloning |
| Lc-TRAF6-R | AGCCTCGGTTTCAAGGGAC | |
| pTurbo-TRAF6-F | CCGGAATTCTGATGGCTTGCATTGACAGCAAT | pTurbo-TRAF6-GFP |
| pTurbo-TRAF6-R | CGCGGATCCCGGTCCCTTGAAACCGAGGCT | |
| qTRAF6-F | GACGGACGGTTGGTAAAGCAG | RT-qPCR |
| qTRAF6-R | CAACTTGTAGCCTGGACGACCC | |
| qβ-actin-F | TTATGAAGGCTATGCCCTGCC | RT-qPCR |
| qβ-actin-R | TGAAGGAGTAGCCACGCTCTGT |

Fig. 1 Multiple alignment of Lc-TRAF6 with TRAF6 in other vertebrates The black arrows represent the RING finger domain,zinc finger domain, coiled-coil domain, and MATH domain respectively
| 常用名Common name | 物种名Scientific name | NCBI序列登录号Accession No. | 序列长度Length/aa | 一致性Identity/% | 相似性Similarity/% |
|---|---|---|---|---|---|
| Grouper | Epinephelus coioides | AGQ45557.1 | 570 | 89 | 94 |
| Fugu | Takifugu rubripes | XP_011608207.2 | 564 | 79 | 87 |
| Rainbow trout | Oncorhynchus mykiss | AVM80404.1 | 551 | 66 | 78 |
| Grass carp | Ctenopharyngodon idella | AGI51678.1 | 542 | 63 | 75 |
| Zebrafish | Danio rerio | NP_001038217.1 | 542 | 61 | 74 |
| channel catfish | Ictalurus punctatus | XP_017313923.1 | 540 | 60 | 74 |
| Chicken | Gallus gallus | XP_015142694.1 | 545 | 52 | 66 |
| Human | Homo sapiens | NP_665802.1 | 522 | 51 | 65 |
| Mouse | Mus musculus | NP_001290202.1 | 530 | 50 | 64 |
Table 2 Comparison of amino acid sequence similarity between Lc-TRAF6 and TRAF6 in other vertebrates
| 常用名Common name | 物种名Scientific name | NCBI序列登录号Accession No. | 序列长度Length/aa | 一致性Identity/% | 相似性Similarity/% |
|---|---|---|---|---|---|
| Grouper | Epinephelus coioides | AGQ45557.1 | 570 | 89 | 94 |
| Fugu | Takifugu rubripes | XP_011608207.2 | 564 | 79 | 87 |
| Rainbow trout | Oncorhynchus mykiss | AVM80404.1 | 551 | 66 | 78 |
| Grass carp | Ctenopharyngodon idella | AGI51678.1 | 542 | 63 | 75 |
| Zebrafish | Danio rerio | NP_001038217.1 | 542 | 61 | 74 |
| channel catfish | Ictalurus punctatus | XP_017313923.1 | 540 | 60 | 74 |
| Chicken | Gallus gallus | XP_015142694.1 | 545 | 52 | 66 |
| Human | Homo sapiens | NP_665802.1 | 522 | 51 | 65 |
| Mouse | Mus musculus | NP_001290202.1 | 530 | 50 | 64 |
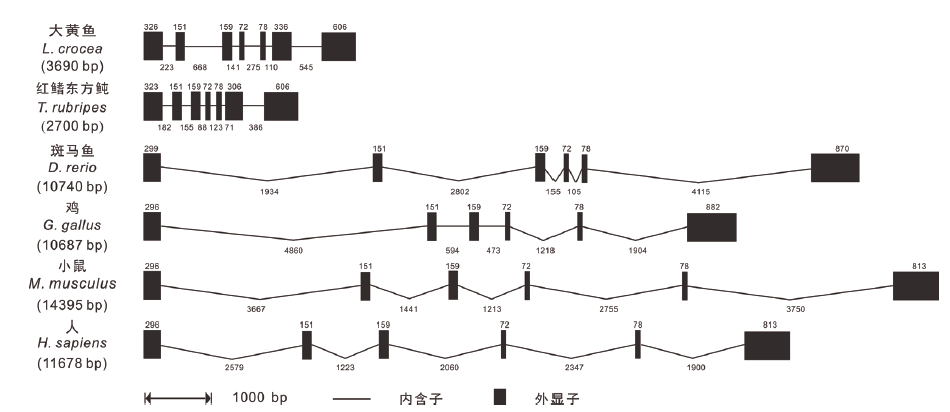
Fig.2 Genomic structure comparison of gene Lc-TRAF6 with TRAF6 in other vertebrates Structure comparison of exons and introns of TRAF6 gene in L. crocea,T. rubripes,D. rerio,G. gallus,M. musculus and H. sapiens. Exons and introns are represented by black boxes and lines respectively,with the length of the exon shown above the black box and the length of the intron shown below the line. The length of the black box and the length of the line is proportional to the length of the sequence. Gene sequences information and their GenBank accession numbers are shown as follows:L. crocea,NC_040018.1(4990344-4997110);T. rubripes,NC_042297.1(13166964-13171483);D. rerio,NC_0071187(48722838-48738718);G. gallus,NC_052536.1(19015766-19036372);M. musculus,NC_000068.8(101508655-101532013);H. sapiens,NC_000011.10(36483769-36510313)
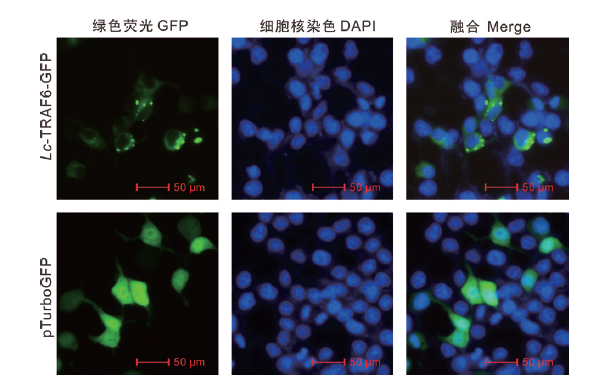
Fig.3 Subcellular localization analysis of Lc-TRAF6 HEK 293T cells were transiently transfected with pTurboGFP and pTurbo-TRAF6-GFP respectively. At 24 h after transfection,cells were stained with DAPI,detected under a confocal microscope and photographed
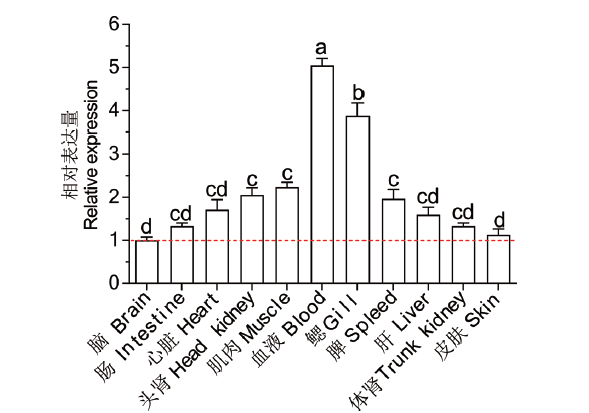
Fig. 4 Tissue expression analysis of Lc-TRAF6 The expression pattern of Lc-TRAF6 mRNA in 11 different tissues/organs of healthy large yellow croaker was detected by quantitative real-time PCR analysis. The mRNA expression level of Lc-TRAF6 in the brain was set as 1-fold,and the mRNA expression level of Lc-TRAF6 in other organs/tissues was recorded as multiple of the expression level in the brain,and the baseline of 1-fold was marked with red dotted line. All data were expressed as mean±SE(n = 6). Different superscripts indicate statistically different results(P < 0.05)and the same superscript indicates no statistical differences between groups
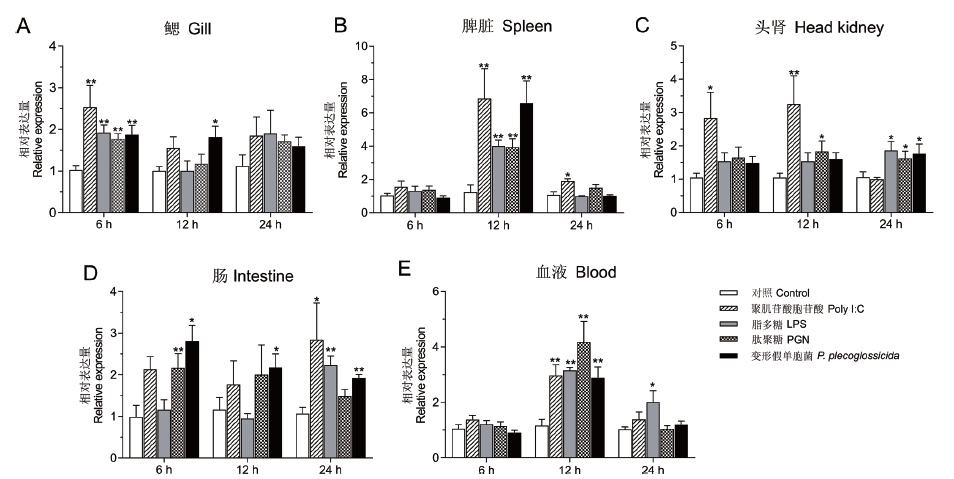
Fig. 5 Expression analysis of Lc-TRAF6 under Poly I:C,LPS,PGN,and P. plecoglossicida stimulation The healthy large yellow croakers was stimulated with Poly I:C,LPS,PGN,and P. plecoglossicida for 6,12,and 24 h(with PBS set as the control group),then the mRNA expression levels of Lc-TRAF6 in gill(A),spleen(B),head kidney(C),intestine(D),and blood(E)were detected by RT-qPCR. The results were normalized to the expression of β-actin and the data were recorded as mean ±SE(n = 6). * P < 0.05,**P < 0.01
| [1] |
Dhillon B, Aleithan F, Abdul-Sater Z, et al. The evolving role of TRAFs in mediating inflammatory responses[J]. Front Immunol, 2019, 10:104.
doi: 10.3389/fimmu.2019.00104 URL |
| [2] |
Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family:scaffold molecules for cytokine receptors, kinases and their regulators[J]. Cell Signal, 2001, 13(6):389-400.
doi: 10.1016/s0898-6568(01)00160-7 pmid: 11384837 |
| [3] |
Xie P. TRAF molecules in cell signaling and in human diseases[J]. J Mol Signal, 2013, 8(1):7.
doi: 10.1186/1750-2187-8-7 URL |
| [4] |
Lalani AI, Zhu SN, Gokhale S, et al. TRAF molecules in inflammation and inflammatory diseases[J]. Curr Pharmacol Rep, 2018, 4(1):64-90.
doi: 10.1007/s40495-017-0117-y pmid: 29527458 |
| [5] |
Zou PF, Tang JC, Li Y, et al. MAVS splicing variants associated with TRAF3 and TRAF6 in NF-κB and IRF3 signaling pathway in large yellow croaker Larimichthys crocea[J]. Dev Comp Immunol, 2021, 121:104076.
doi: 10.1016/j.dci.2021.104076 URL |
| [6] |
Zotti T, Vito P, Stilo R. The seventh ring:exploring TRAF7 functions[J]. J Cell Physiol, 2012, 227(3):1280-1284.
doi: 10.1002/jcp.24011 URL |
| [7] |
Arch RH, Gedrich RW, Thompson CB. Tumor necrosis factor receptor-associated factors(TRAFs)—a family of adapter proteins that regulates life and death[J]. Genes Dev, 1998, 12(18):2821-2830.
doi: 10.1101/gad.12.18.2821 URL |
| [8] | 施伟梅, 杨凯, 李林福, 等. IRE1/TRAF2诱导细胞凋亡的分子机制[J]. 基因组学与应用生物学, 2018, 37(6):2579-2582. |
| Shi WM, Yang K, Li LF, et al. Molecular mechanism of cell apoptosis induced by IRE1/TRAF2[J]. Genom Appl Biol, 2018, 37(6):2579-2582. | |
| [9] |
Li FB, Li Y, Liang HC, et al. HECTD3 mediates TRAF3 polyubiquitination and type I interferon induction during bacterial infection[J]. J Clin Invest, 2018, 128(9):4148-4162.
doi: 10.1172/JCI120406 URL |
| [10] |
Martinez-Forero I, Rouzaut A, Palazon A, et al. Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation[J]. Clin Cancer Res, 2009, 15(22):6751-6757.
doi: 10.1158/1078-0432.CCR-09-1225 pmid: 19887490 |
| [11] |
Wang XW, Yang J, Han L, et al. TRAF5-mediated Lys-63-linked polyubiquitination plays an essential role in positive regulation of RORγt in promoting IL-17A expression[J]. J Biol Chem, 2015, 290(48):29086-29094.
doi: 10.1074/jbc.M115.664573 URL |
| [12] |
Li YW, Li X, Xiao XX, et al. Molecular characterization and functional analysis of TRAF6 in orange-spotted grouper(Epinephelus coioides)[J]. Dev Comp Immunol, 2014, 44(1):217-225.
doi: 10.1016/j.dci.2013.12.011 URL |
| [13] |
Darnay BG, Ni J, Moore PA, et al. Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor(TRAF)6 and NF-kappaB-inducing kinase. Identification of a novel TRAF6 interaction motif[J]. J Biol Chem, 1999, 274(12):7724-7731.
doi: 10.1074/jbc.274.12.7724 pmid: 10075662 |
| [14] |
Pollet I, Opina CJ, Zimmerman C, et al. Bacterial lipopolysaccharide directly induces angiogenesis through TRAF6-mediated activation of NF-kappaB and c-Jun N-terminal kinase[J]. Blood, 2003, 102(5):1740-1742.
doi: 10.1182/blood-2003-01-0288 URL |
| [15] |
Kanamori M, Kai C, Hayashizaki Y, et al. NF-kappaB activator Act1 associates with IL-1/Toll pathway adaptor molecule TRAF6[J]. FEBS Lett, 2002, 532(1/2):241-246.
doi: 10.1016/S0014-5793(02)03688-8 URL |
| [16] |
Kang LS, Wang LP, Wu CW, et al. Molecular characterization and expression analysis of tumor necrosis factor receptor-associated factors 3 and 6 in large yellow croaker(Larimichthys crocea)[J]. Fish Shellfish Immunol, 2018, 82:27-31.
doi: 10.1016/j.fsi.2018.07.051 URL |
| [17] | Wang JJ, Wu XJ, Jiang MY, et al. Mechanism by which TRAF6 participates in the immune regulation of autoimmune diseases and cancer[J]. Biomed Res Int, 2020, 2020:4607197. |
| [18] |
Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5):637-650.
doi: 10.1016/j.immuni.2011.05.006 pmid: 21616434 |
| [19] |
Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity:update on Toll-like receptors[J]. Nat Immunol, 2010, 11(5):373-384.
doi: 10.1038/ni.1863 pmid: 20404851 |
| [20] |
Deng L, Wang C, Spencer E, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain[J]. Cell, 2000, 103(2):351-361.
doi: 10.1016/s0092-8674(00)00126-4 pmid: 11057907 |
| [21] |
Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer[J]. Trends Cell Biol, 2009, 19(8):404-413.
doi: 10.1016/j.tcb.2009.05.006 pmid: 19648011 |
| [22] |
Konno H, Yamamoto T, Yamazaki K, et al. TRAF6 establishes innate immune responses by activating NF-kappaB and IRF7 upon sensing cytosolic viral RNA and DNA[J]. PLoS One, 2009, 4(5):e5674.
doi: 10.1371/journal.pone.0005674 URL |
| [23] |
Peng F, Jin SS, Chen ZY, et al. TRIF-mediated antiviral signaling is differentially regulated by TRAF2 and TRAF6 in black carp[J]. Dev Comp Immunol, 2021, 121:104073.
doi: 10.1016/j.dci.2021.104073 URL |
| [24] |
Wang PF, Li FX, Zhao C, et al. Molecular characterization and functional analysis of TRAF6 in the spotted sea bass(Lateolabrax maculatus)[J]. Fish Shellfish Immunol, 2020, 105:233-243.
doi: 10.1016/j.fsi.2020.06.048 URL |
| [25] |
Li YP, Mao YX, Yu NL, et al. Grass carp(Ctenopharyngodon idellus)TRAF6 up-regulates IFN1 expression by activating IRF5[J]. Dev Comp Immunol, 2020, 102:103475.
doi: 10.1016/j.dci.2019.103475 URL |
| [26] |
Zhang J, Zhu YC, Chen Z, et al. Molecular cloning and expression analysis of MyD88 and TRAF6 in Qihe crucian carp Carassius auratus[J]. Fish Shellfish Immunol, 2019, 87:829-838.
doi: 10.1016/j.fsi.2019.02.034 URL |
| [27] |
Jang JH, Kim H, Cho JH. Molecular cloning and functional characterization of TRAF6 and TAK1 in rainbow trout, Oncorhynchus mykiss[J]. Fish Shellfish Immunol, 2019, 84:927-936.
doi: 10.1016/j.fsi.2018.11.002 URL |
| [28] | 刘家富, 韩坤煌. 我国大黄鱼产业的发展现状与对策[J]. 福建水产, 2011, 33(5):4-8. |
| Liu JF, Han KH. Current development situation and countermeasure of large yellow crocker industry in China[J]. J Fujian Fish, 2011, 33(5):4-8. | |
| [29] | 徐春霞. 网箱养殖大黄鱼内脏白点病病原菌分离鉴定及致病性研究[J]. 水产科学, 2021, 40(5):670-678. |
| Xu CX. Isolation, identification and pathogenicity of white-nodules disease in internal organs of large yellow croaker pseudosciaena crocea in a sea cage[J]. Fish Sci, 2021, 40(5):670-678. | |
| [30] | 游剑涛. 大黄鱼烂鳃病病原菌的分离鉴定及药敏分析[J]. 渔业研究, 2018, 40(6):425-433. |
| You JT. Isolation, identification and antibiotic sensitivity analysis of bacterial pathogen from Larimichthys crocea with gill-rot disease[J]. J Fish Res, 2018, 40(6):425-433. | |
| [31] | 施慧, 陈卓, 丁慧昕, 等. 养殖大黄鱼一种黏孢子虫病的组织病理学及检测方法初探[J]. 中国水产科学, 2019, 26(1):203-213. |
| Shi H, Chen Z, Ding HX, et al. Preliminary study on the histopathology and detection methods of a Myxosporea parasite that causes white-gill disease in cultured Larimichthys crocea[J]. J Fish Res, 2019, 26(1):203-213. | |
| [32] | 吴静. 饥饿对大黄鱼体内虹彩病毒载量及相关免疫反应影响的初步研究[D]. 舟山: 浙江海洋大学, 2020. |
| Wu J. A preliminary study on the effects of starvation on the iridovirus load and related immune responses in large yellow croaker[D]. Zhoushan: Zhejiang Ocean University, 2020. | |
| [33] |
Shi JH, Sun SC. Tumor necrosis factor receptor-associated factor regulation of nuclear factor κB and mitogen-activated protein kinase pathways[J]. Front Immunol, 2018, 9:1849.
doi: 10.3389/fimmu.2018.01849 URL |
| [34] |
Chen H, Xiao J, Li J, et al. TRAF2 of black carp upregulates MAVS-mediated antiviral signaling during innate immune response[J]. Fish Shellfish Immunol, 2017, 71:1-9.
doi: 10.1016/j.fsi.2017.09.069 URL |
| [35] |
Zhao F, Li YW, Pan HJ, et al. Grass carp(Ctenopharyngodon idella)TRAF6 and TAK1:molecular cloning and expression analysis after Ichthyophthirius multifiliis infection[J]. Fish Shellfish Immunol, 2013, 34(6):1514-1523.
doi: 10.1016/j.fsi.2013.03.003 URL |
| [36] |
Wang ZW, Huang Y, Li Y, et al. Biological characterization, expression, and functional analysis of tumor necrosis factor receptor-associated factor 6 in Nile tilapia(Oreochromis niloticus)[J]. Fish Shellfish Immunol, 2018, 80:497-504.
doi: 10.1016/j.fsi.2018.06.036 URL |
| [37] |
Jiang S, Xiao J, Li J, et al. Characterization of the black carp TRAF6 signaling molecule in innate immune defense[J]. Fish Shellfish Immunol, 2017, 67:147-158.
doi: 10.1016/j.fsi.2017.06.011 URL |
| [38] |
Wang PF, Li FX, Zhao C, et al. Molecular characterization and functional analysis of TRAF6 in the spotted sea bass(Lateolabrax maculatus)[J]. Fish Shellfish Immunol, 2020, 105:233-243.
doi: 10.1016/j.fsi.2020.06.048 URL |
| [39] | 罗智文, 董志祥, 林连兵, 等. 鱼类重要免疫器官抗菌机制的研究进展[J]. 水产科学, 2021, 40(4):624-634. |
| Luo ZW, Dong ZX, Lin LB, et al. Advances on immunological mechanisms of important immune organs against pathogenic microorganisms in fish:a review[J]. Fish Sci, 2021, 40(4):624-634. | |
| [40] | 姜红烨, 黄艳, 余新炳. 鱼的黏膜免疫研究进展[J]. 热带医学杂志, 2015, 15(8):1150-1153. |
| Jiang HY, Huang Y, Yu XB. Research progress in mucosal immunity of fish[J]. J Trop Med, 2015, 15(8):1150-1153. | |
| [41] | 贾生美. 鲤鱼TLR通路中TRAF6的基因克隆、鉴定及差异表达分析[D]. 长春: 吉林大学, 2014. |
| Jia SM. Cloning, characterization and expression analysis of common carp TRAF6[D]. Changchun: Jilin University, 2014. |
| [1] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [2] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [3] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [4] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [5] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [6] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [7] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [8] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [9] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [10] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [11] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [12] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [13] | YOU Chui-huai, XIE Jin-jin, ZHANG Ting, CUI Tian-zhen, SUN Xin-lu, ZANG Shou-jian, WU Yi-ning, SUN Meng-yao, QUE You-xiong, SU Ya-chun. Identification of the Lipoxygenase Gene GeLOX1 and Expression Analysis Under Low Temperature Stress in Gelsmium elegans [J]. Biotechnology Bulletin, 2023, 39(11): 318-327. |
| [14] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [15] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||