








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (11): 270-282.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0530
Previous Articles Next Articles
CHEN Chu-yi( ), YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran(
), YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran( )
)
Received:2023-06-05
Online:2023-11-26
Published:2023-12-20
Contact:
YUE Li-ran
E-mail:chen629@qq.com;ms_yueliran@163.com
CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment[J]. Biotechnology Bulletin, 2023, 39(11): 270-282.
| 基因 Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CmEF1α | TTTTGGTATCTGGTCCTGGAG | CCATTCAAGCGACAGACTCA |
| CnZF-HD1 | CCGCGTGTAACTGTCATCGT | GTGACGTGGATGGTAACGCT |
| CnZF-HD2 | GTCACCACCATAGCAACTACCA | CCACCATAACCAGCACCATCT |
| CnZF-HD3 | GAAGAGAAGAACCACCACCCT | AGTTTCCACCACCCTTCTATGG |
| CnZF-HD4 | ACCACCTTACACCGCCTTTC | AAAGGACACGGGTAGTCCAC |
| CnZF-HD5 | ATGCAGTAGACGGATGCAGAG | CTCTACGATGGAAATTCCTGTGGC |
| CnZF-HD6 | CCCTCACCTTCAGCGTCATT | TTCTCCTTTTGTTCGGGCGT |
| CnZF-HD7 | AGTTTCCACCGGAGAGAGGT | CATTTGCCCCACCAAATGCC |
| CnZF-HD8 | TGGAGAAGAAGGTACACCCGA | TGATGGCTGATGCTCGGTTT |
| CnZF-HD9 | TGGCCTCACTACCATCAACC | CAGTGTGTAGCGGCATGAAC |
| CnZF-HD10 | CTTTGGATGGGTGTAGGGAGTT | TCCTCCTGTGGAAACTCTGGTG |
| CnZF-HD11 | TACTACCCTTCAGCCCCACA | CATCTTCCATCCCACACGCT |
| CnZF-HD12 | CCTTCAGCATTTGTGCCAGT | CATCCGGTCCTTCTGAGCTTT |
| CnZF-HD13 | CTTCTGTAAATGCCCCACCC | TTCTCTTGGTATCCTCCACCC |
| CnZF-HD14 | CACCAACCATTGCTACCTCCT | AACTCCCCGCAACCATCAAG |
| CnZF-HD15 | CCACCCCATTCCCAATCAATC | TTTGAATGAAGATGGCTCCGCC |
| CnZF-HD16 | TGGCTGTCACCGGAGTTTTC | ATCACCGGCACCGTTTTTGG |
| CnZF-HD17 | TCCACCATGCAAGTGTACC | TCCCTCTTCTTATCCCAATCCC |
| CnZF-HD18 | TGGGAGACGAGAAAAAGAGTCC | CACCTTTAGTGCCCTCCTC |
| CnZF-HD19 | TGGTTGCGGGGAGTTTATGT | TGTGAGGTGGTGGTGGATAG |
Table 1 RT-qPCR primers
| 基因 Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CmEF1α | TTTTGGTATCTGGTCCTGGAG | CCATTCAAGCGACAGACTCA |
| CnZF-HD1 | CCGCGTGTAACTGTCATCGT | GTGACGTGGATGGTAACGCT |
| CnZF-HD2 | GTCACCACCATAGCAACTACCA | CCACCATAACCAGCACCATCT |
| CnZF-HD3 | GAAGAGAAGAACCACCACCCT | AGTTTCCACCACCCTTCTATGG |
| CnZF-HD4 | ACCACCTTACACCGCCTTTC | AAAGGACACGGGTAGTCCAC |
| CnZF-HD5 | ATGCAGTAGACGGATGCAGAG | CTCTACGATGGAAATTCCTGTGGC |
| CnZF-HD6 | CCCTCACCTTCAGCGTCATT | TTCTCCTTTTGTTCGGGCGT |
| CnZF-HD7 | AGTTTCCACCGGAGAGAGGT | CATTTGCCCCACCAAATGCC |
| CnZF-HD8 | TGGAGAAGAAGGTACACCCGA | TGATGGCTGATGCTCGGTTT |
| CnZF-HD9 | TGGCCTCACTACCATCAACC | CAGTGTGTAGCGGCATGAAC |
| CnZF-HD10 | CTTTGGATGGGTGTAGGGAGTT | TCCTCCTGTGGAAACTCTGGTG |
| CnZF-HD11 | TACTACCCTTCAGCCCCACA | CATCTTCCATCCCACACGCT |
| CnZF-HD12 | CCTTCAGCATTTGTGCCAGT | CATCCGGTCCTTCTGAGCTTT |
| CnZF-HD13 | CTTCTGTAAATGCCCCACCC | TTCTCTTGGTATCCTCCACCC |
| CnZF-HD14 | CACCAACCATTGCTACCTCCT | AACTCCCCGCAACCATCAAG |
| CnZF-HD15 | CCACCCCATTCCCAATCAATC | TTTGAATGAAGATGGCTCCGCC |
| CnZF-HD16 | TGGCTGTCACCGGAGTTTTC | ATCACCGGCACCGTTTTTGG |
| CnZF-HD17 | TCCACCATGCAAGTGTACC | TCCCTCTTCTTATCCCAATCCC |
| CnZF-HD18 | TGGGAGACGAGAAAAAGAGTCC | CACCTTTAGTGCCCTCCTC |
| CnZF-HD19 | TGGTTGCGGGGAGTTTATGT | TGTGAGGTGGTGGTGGATAG |
| 基因ID Gene ID | 基因名称 Gene name | 氨基酸 Number of amino acids/aa | 分子量 Molecular weight/Da | 等电点 pI | 亚细胞定位 Predicted location | 磷酸化位点数目Phosphorylation site | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 丝氨酸 Ser | 苏氨酸 Thr | 酪氨酸 Tyr | 总数 Total number | |||||||
| CHR00000129 | CnZF-HD1 | 239 | 26 293.14 | 8.70 | 细胞核 Nucleus | 16 | 12 | 1 | 29 | |
| CHR00004454 | CnZF-HD2 | 324 | 36 580.21 | 7.14 | 细胞核 Nucleus | 11 | 7 | 3 | 21 | |
| CHR00008752 | CnZF-HD3 | 95 | 10 527.72 | 5.77 | 细胞质 Cytoplasm | 7 | 4 | 1 | 12 | |
| CHR00009701 | CnZF-HD4 | 337 | 37 541.18 | 6.80 | 细胞核 Nucleus | 24 | 12 | 4 | 40 | |
| CHR00010377 | CnZF-HD5 | 94 | 10 259.57 | 7.60 | 线粒体 Mitochondria | 9 | 6 | 0 | 15 | |
| CHR00019161 | CnZF-HD6 | 267 | 29 598.21 | 6.83 | 细胞核 Nucleus | 15 | 15 | 4 | 34 | |
| CHR00030290 | CnZF-HD7 | 434 | 49 475.83 | 9.57 | 叶绿体 Chloroplast | 24 | 19 | 3 | 46 | |
| CHR00046942 | CnZF-HD8 | 263 | 30 175.71 | 8.21 | 细胞核 Nucleus | 15 | 11 | 3 | 29 | |
| CHR00047218 | CnZF-HD9 | 258 | 28 269.12 | 9.27 | 细胞核 Nucleus | 17 | 4 | 3 | 24 | |
| CHR00048774 | CnZF-HD10 | 85 | 9 282.57 | 7.67 | 叶绿体和线粒体 Chloroplast and Mitochondria | 5 | 1 | 1 | 7 | |
| CHR00051473 | CnZF-HD11 | 349 | 38 055.30 | 7.79 | 细胞核 Nucleus | 22 | 14 | 1 | 37 | |
| CHR00055582 | CnZF-HD12 | 191 | 21 238.01 | 9.71 | 细胞核 Nucleus | 12 | 5 | 1 | 18 | |
| CHR00058475 | CnZF-HD13 | 289 | 32 048.70 | 8.49 | 细胞核 Nucleus | 23 | 11 | 2 | 36 | |
| CHR00061161 | CnZF-HD14 | 145 | 16 331.64 | 7.08 | 叶绿体 Chloroplast | 14 | 3 | 0 | 17 | |
| CHR00077153 | CnZF-HD15 | 218 | 23 736.40 | 6.19 | 细胞核 Nucleus | 12 | 18 | 1 | 31 | |
| CHR00081707 | CnZF-HD16 | 123 | 13 516.44 | 9.87 | 线粒体 Mitochondria | 9 | 4 | 1 | 14 | |
| CHR00082507 | CnZF-HD17 | 309 | 33 442.00 | 6.63 | 细胞核 Nucleus | 17 | 24 | 2 | 43 | |
| CHR00084858 | CnZF-HD18 | 91 | 10 311.30 | 4.69 | 细胞核 Nucleus | 7 | 1 | 2 | 10 | |
| CHR00091694 | CnZF-HD19 | 173 | 19 332.72 | 6.27 | 叶绿体 Chloroplast | 16 | 7 | 1 | 24 | |
Table 2 Physicochemical properties of CnZF-HD family proteins
| 基因ID Gene ID | 基因名称 Gene name | 氨基酸 Number of amino acids/aa | 分子量 Molecular weight/Da | 等电点 pI | 亚细胞定位 Predicted location | 磷酸化位点数目Phosphorylation site | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 丝氨酸 Ser | 苏氨酸 Thr | 酪氨酸 Tyr | 总数 Total number | |||||||
| CHR00000129 | CnZF-HD1 | 239 | 26 293.14 | 8.70 | 细胞核 Nucleus | 16 | 12 | 1 | 29 | |
| CHR00004454 | CnZF-HD2 | 324 | 36 580.21 | 7.14 | 细胞核 Nucleus | 11 | 7 | 3 | 21 | |
| CHR00008752 | CnZF-HD3 | 95 | 10 527.72 | 5.77 | 细胞质 Cytoplasm | 7 | 4 | 1 | 12 | |
| CHR00009701 | CnZF-HD4 | 337 | 37 541.18 | 6.80 | 细胞核 Nucleus | 24 | 12 | 4 | 40 | |
| CHR00010377 | CnZF-HD5 | 94 | 10 259.57 | 7.60 | 线粒体 Mitochondria | 9 | 6 | 0 | 15 | |
| CHR00019161 | CnZF-HD6 | 267 | 29 598.21 | 6.83 | 细胞核 Nucleus | 15 | 15 | 4 | 34 | |
| CHR00030290 | CnZF-HD7 | 434 | 49 475.83 | 9.57 | 叶绿体 Chloroplast | 24 | 19 | 3 | 46 | |
| CHR00046942 | CnZF-HD8 | 263 | 30 175.71 | 8.21 | 细胞核 Nucleus | 15 | 11 | 3 | 29 | |
| CHR00047218 | CnZF-HD9 | 258 | 28 269.12 | 9.27 | 细胞核 Nucleus | 17 | 4 | 3 | 24 | |
| CHR00048774 | CnZF-HD10 | 85 | 9 282.57 | 7.67 | 叶绿体和线粒体 Chloroplast and Mitochondria | 5 | 1 | 1 | 7 | |
| CHR00051473 | CnZF-HD11 | 349 | 38 055.30 | 7.79 | 细胞核 Nucleus | 22 | 14 | 1 | 37 | |
| CHR00055582 | CnZF-HD12 | 191 | 21 238.01 | 9.71 | 细胞核 Nucleus | 12 | 5 | 1 | 18 | |
| CHR00058475 | CnZF-HD13 | 289 | 32 048.70 | 8.49 | 细胞核 Nucleus | 23 | 11 | 2 | 36 | |
| CHR00061161 | CnZF-HD14 | 145 | 16 331.64 | 7.08 | 叶绿体 Chloroplast | 14 | 3 | 0 | 17 | |
| CHR00077153 | CnZF-HD15 | 218 | 23 736.40 | 6.19 | 细胞核 Nucleus | 12 | 18 | 1 | 31 | |
| CHR00081707 | CnZF-HD16 | 123 | 13 516.44 | 9.87 | 线粒体 Mitochondria | 9 | 4 | 1 | 14 | |
| CHR00082507 | CnZF-HD17 | 309 | 33 442.00 | 6.63 | 细胞核 Nucleus | 17 | 24 | 2 | 43 | |
| CHR00084858 | CnZF-HD18 | 91 | 10 311.30 | 4.69 | 细胞核 Nucleus | 7 | 1 | 2 | 10 | |
| CHR00091694 | CnZF-HD19 | 173 | 19 332.72 | 6.27 | 叶绿体 Chloroplast | 16 | 7 | 1 | 24 | |
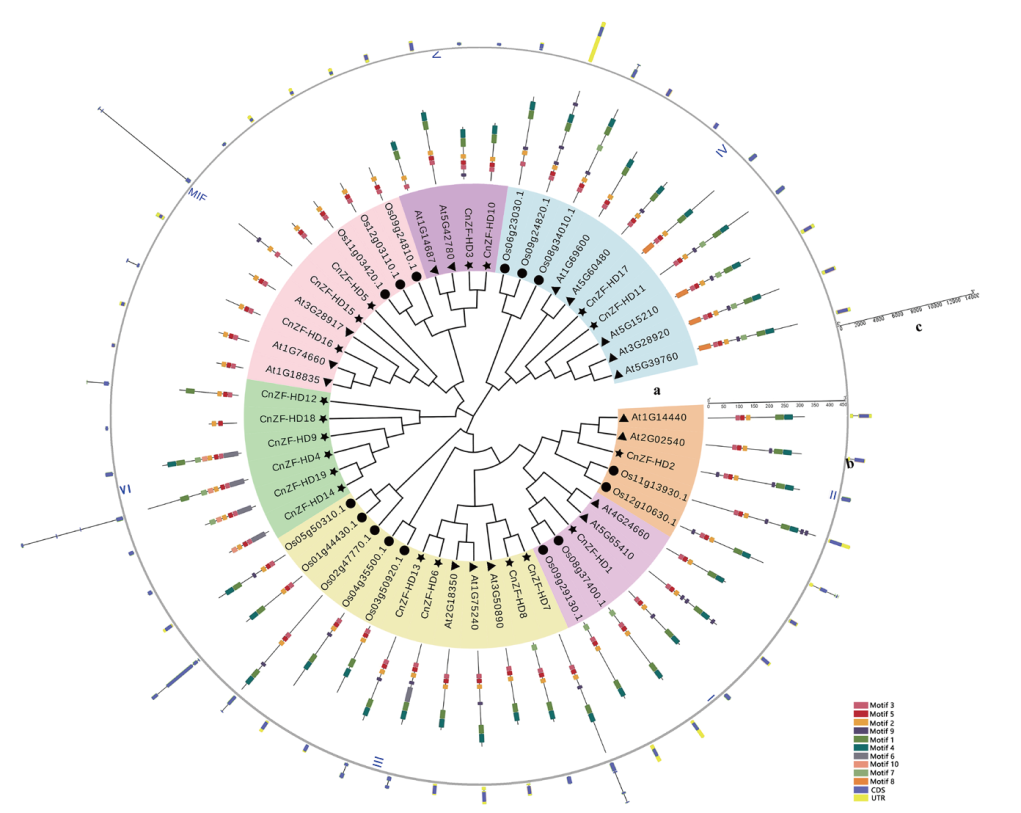
Fig. 2 Phylogenetic analysis (a), conservative motif (b), and gene structure (c) of ZF-HD genes in A. thaliana,O. sativa,and C. nankingense AT: Arabidopsis thaliana; Os: Oryza sativa; Cn: Chrysanthemum nankingense

Fig. 3 Analysis of cis-acting elements of the promoter of CnZF-HD family genes A: Number of different cis-acting elements in the CnZF-HD gene. B: Number of cis-acting elements in each category. C: Ratio of different cis-acting elements in each category
| 基因 Gene | 处理时间 Treatment time/h | |||||
|---|---|---|---|---|---|---|
| 0 | 2 | 6 | 12 | 24 | ||
| CnZF-HD1 | (1.00±0.00)d | (1.52±0.11)c | (4.02±0.03)b | (4.36±0.02)b | (5.14±0.05)a | |
| CnZF-HD2 | (1.00±0.00)d | (1.76±0.13)c | (3.05±0.02)a | (2.15±0.04)b | (1.53±0.04)c | |
| CnZF-HD3 | (1.00±0.00)b | (2.12±0.03)a | (1.34±0.07)b | (1.24±0.05)b | (1.13±0.04)b | |
| CnZF-HD4 | (1.00±0.00)b | (1.05±0.02)b | (1.87±0.11)a | (0.84±0.09)c | (0.73±0.11)c | |
| CnZF-HD5 | (1.00±0.00)d | (1.48±0.11)c | (3.41±0.06)b | (3.97±0.07)a | (1.41±0.09)c | |
| CnZF-HD6 | (1.00±0.00)c | (1.28±0.03)b | (1.38±0.10)b | (2.13±0.05)a | (0.94±0.07)c | |
| CnZF-HD7 | (1.00±0.00)c | (2.14±0.14)a | (1.35±0.12)b | (1.19±0.11)c | (0.58±0.08)d | |
| CnZF-HD8 | (1.00±0.00)c | (2.08±0.09)a | (1.28±0.08)b | (1.2±0.06)b | (0.62±0.07)d | |
| CnZF-HD9 | (1.00±0.00)c | (1.32±0.09)b | (1.56±0.07)a | (0.84±0.11)d | (0.71±0.12)d | |
| CnZF-HD10 | (1.00±0.00)c | (1.88±0.06)a | (1.34±0.12)b | (1.21±0.13)b | (1.08±0.12)c | |
| CnZF-HD11 | (1.00±0.00)c | (1.21±0.05)b | (1.53±0.04)a | (1.37±0.14)b | (1.14±0.11)c | |
| CnZF-HD12 | (1.00±0.00)c | (1.28±0.05)b | (2.19±0.09)a | (0.78±0.06)d | (0.68±0.07)d | |
| CnZF-HD13 | (1.00±0.00)c | (1.24±0.04)b | (1.29±0.14)b | (1.75±0.03)a | (0.63±0.02)d | |
| CnZF-HD14 | (1.00±0.00)c | (1.89±0.05)b | (2.17±0.10)a | (0.81±0.11)b | (0.73±0.13)d | |
| CnZF-HD15 | (1.00±0.00)d | (1.52±0.13)c | (3.49±0.07)b | (4.08±0.08)a | (1.39±0.08)c | |
| CnZF-HD16 | (1.00±0.00)c | (1.53±0.13)b | (3.53±0.08)a | (3.78±0.11)a | (1.43±0.11)b | |
| CnZF-HD17 | (1.00±0.00)b | (1.12±0.11)b | (1.38±0.05)a | (1.26±0.04)a | (0.72±0.07)c | |
| CnZF-HD18 | (1.00±0.00)c | (1.32±0.06)b | (2.04±0.07)a | (1.08±0.11)c | (0.74±0.09)d | |
| CnZF-HD19 | (1.00±0.00)b | (2.01±0.07)a | (2.17±0.08)a | (0.83±0.08)c | (0.74±0.06)c | |
Table 3 Relative expressions of CnZF-HDs under drought treatment
| 基因 Gene | 处理时间 Treatment time/h | |||||
|---|---|---|---|---|---|---|
| 0 | 2 | 6 | 12 | 24 | ||
| CnZF-HD1 | (1.00±0.00)d | (1.52±0.11)c | (4.02±0.03)b | (4.36±0.02)b | (5.14±0.05)a | |
| CnZF-HD2 | (1.00±0.00)d | (1.76±0.13)c | (3.05±0.02)a | (2.15±0.04)b | (1.53±0.04)c | |
| CnZF-HD3 | (1.00±0.00)b | (2.12±0.03)a | (1.34±0.07)b | (1.24±0.05)b | (1.13±0.04)b | |
| CnZF-HD4 | (1.00±0.00)b | (1.05±0.02)b | (1.87±0.11)a | (0.84±0.09)c | (0.73±0.11)c | |
| CnZF-HD5 | (1.00±0.00)d | (1.48±0.11)c | (3.41±0.06)b | (3.97±0.07)a | (1.41±0.09)c | |
| CnZF-HD6 | (1.00±0.00)c | (1.28±0.03)b | (1.38±0.10)b | (2.13±0.05)a | (0.94±0.07)c | |
| CnZF-HD7 | (1.00±0.00)c | (2.14±0.14)a | (1.35±0.12)b | (1.19±0.11)c | (0.58±0.08)d | |
| CnZF-HD8 | (1.00±0.00)c | (2.08±0.09)a | (1.28±0.08)b | (1.2±0.06)b | (0.62±0.07)d | |
| CnZF-HD9 | (1.00±0.00)c | (1.32±0.09)b | (1.56±0.07)a | (0.84±0.11)d | (0.71±0.12)d | |
| CnZF-HD10 | (1.00±0.00)c | (1.88±0.06)a | (1.34±0.12)b | (1.21±0.13)b | (1.08±0.12)c | |
| CnZF-HD11 | (1.00±0.00)c | (1.21±0.05)b | (1.53±0.04)a | (1.37±0.14)b | (1.14±0.11)c | |
| CnZF-HD12 | (1.00±0.00)c | (1.28±0.05)b | (2.19±0.09)a | (0.78±0.06)d | (0.68±0.07)d | |
| CnZF-HD13 | (1.00±0.00)c | (1.24±0.04)b | (1.29±0.14)b | (1.75±0.03)a | (0.63±0.02)d | |
| CnZF-HD14 | (1.00±0.00)c | (1.89±0.05)b | (2.17±0.10)a | (0.81±0.11)b | (0.73±0.13)d | |
| CnZF-HD15 | (1.00±0.00)d | (1.52±0.13)c | (3.49±0.07)b | (4.08±0.08)a | (1.39±0.08)c | |
| CnZF-HD16 | (1.00±0.00)c | (1.53±0.13)b | (3.53±0.08)a | (3.78±0.11)a | (1.43±0.11)b | |
| CnZF-HD17 | (1.00±0.00)b | (1.12±0.11)b | (1.38±0.05)a | (1.26±0.04)a | (0.72±0.07)c | |
| CnZF-HD18 | (1.00±0.00)c | (1.32±0.06)b | (2.04±0.07)a | (1.08±0.11)c | (0.74±0.09)d | |
| CnZF-HD19 | (1.00±0.00)b | (2.01±0.07)a | (2.17±0.08)a | (0.83±0.08)c | (0.74±0.06)c | |
| 基因 Gene | 处理时间 Treatment time/h | |||||
|---|---|---|---|---|---|---|
| 0 | 2 | 6 | 12 | 24 | ||
| CnZF-HD1 | (1.00±0.00)a | (0.45±0.10)d | (0.62±0.07)c | (0.43±0.02)d | (0.82±0.05)b | |
| CnZF-HD2 | (1.00±0.00)a | (0.53±0.16)c | (0.72±0.04)b | (0.47±0.02)c | (0.59±0.02)c | |
| CnZF-HD3 | (1.00±0.00)b | (0.97±0.03)b | (1.02±0.10)a | (0.52±0.04)c | (0.08±0.04)d | |
| CnZF-HD4 | (1.00±0.00)b | (1.35±0.02)a | (1.12±0.10)b | (0.84±0.16)c | (0.53±0.11)d | |
| CnZF-HD5 | (1.00±0.00)a | (0.58±0.13)c | (1.11±0.05)a | (0.87±0.07)b | (0.42±0.13)c | |
| CnZF-HD6 | (1.00±0.00)a | (0.62±0.04)b | (0.36±0.11)c | (0.13±0.07)d | (0.01±0.05)d | |
| CnZF-HD7 | (1.00±0.00)a | (0.74±0.13)b | (0.57±0.14)b | (0.32±0.12)c | (0.19±0.05)d | |
| CnZF-HD8 | (1.00±0.00)a | (0.58±0.12)c | (0.71±0.05)b | (0.47±0.08)c | (0.25±0.07)d | |
| CnZF-HD9 | (1.00±0.00)b | (1.28±0.08)a | (0.96±0.09)b | (0.81±0.13)c | (0.72±0.12)c | |
| CnZF-HD10 | (1.00±0.00)b | (0.87±0.08)c | (1.39±0.11)a | (0.27±0.15)e | (0.54±0.13)d | |
| CnZF-HD11 | (1.00±0.00)a | (0.77±0.07)b | (0.46±0.06)c | (0.59±0.14)c | (0.48±0.10)c | |
| CnZF-HD12 | (1.00±0.00)a | (0.92±0.05)b | (1.09±0.12)a | (0.89±0.06)b | (0.25±0.12)c | |
| CnZF-HD13 | (1.00±0.00)a | (0.64±0.04)b | (0.42±0.16)c | (0.27±0.08)c | (0.15±0.02)d | |
| CnZF-HD14 | (1.00±0.00)b | (1.12±0.05)a | (0.98±0.11)b | (0.65±0.14)c | (0.34±0.13)d | |
| CnZF-HD15 | (1.00±0.00)b | (0.77±0.13)c | (1.42±0.06)a | (1.09±0.08)b | (0.65±0.10)c | |
| CnZF-HD16 | (1.00±0.00)a | (0.60±0.15)b | (1.06±0.09)a | (0.95±0.11)a | (0.48±0.10)c | |
| CnZF-HD17 | (1.00±0.00)a | (0.84±0.11)b | (0.56±0.01)c | (0.62±0.14)c | (0.47±0.07)d | |
| CnZF-HD18 | (1.00±0.00)a | (0.88±0.06)b | (1.02±0.07)a | (0.56±0.15)c | (0.46±0.15)c | |
| CnZF-HD19 | (1.00±0.00)c | (1.77±0.09)a | (1.46±0.07)b | (0.70±0.08)d | (0.56±0.07)d | |
Table 4 Relative expressions of CnZF-HDs in response to ABA treatment
| 基因 Gene | 处理时间 Treatment time/h | |||||
|---|---|---|---|---|---|---|
| 0 | 2 | 6 | 12 | 24 | ||
| CnZF-HD1 | (1.00±0.00)a | (0.45±0.10)d | (0.62±0.07)c | (0.43±0.02)d | (0.82±0.05)b | |
| CnZF-HD2 | (1.00±0.00)a | (0.53±0.16)c | (0.72±0.04)b | (0.47±0.02)c | (0.59±0.02)c | |
| CnZF-HD3 | (1.00±0.00)b | (0.97±0.03)b | (1.02±0.10)a | (0.52±0.04)c | (0.08±0.04)d | |
| CnZF-HD4 | (1.00±0.00)b | (1.35±0.02)a | (1.12±0.10)b | (0.84±0.16)c | (0.53±0.11)d | |
| CnZF-HD5 | (1.00±0.00)a | (0.58±0.13)c | (1.11±0.05)a | (0.87±0.07)b | (0.42±0.13)c | |
| CnZF-HD6 | (1.00±0.00)a | (0.62±0.04)b | (0.36±0.11)c | (0.13±0.07)d | (0.01±0.05)d | |
| CnZF-HD7 | (1.00±0.00)a | (0.74±0.13)b | (0.57±0.14)b | (0.32±0.12)c | (0.19±0.05)d | |
| CnZF-HD8 | (1.00±0.00)a | (0.58±0.12)c | (0.71±0.05)b | (0.47±0.08)c | (0.25±0.07)d | |
| CnZF-HD9 | (1.00±0.00)b | (1.28±0.08)a | (0.96±0.09)b | (0.81±0.13)c | (0.72±0.12)c | |
| CnZF-HD10 | (1.00±0.00)b | (0.87±0.08)c | (1.39±0.11)a | (0.27±0.15)e | (0.54±0.13)d | |
| CnZF-HD11 | (1.00±0.00)a | (0.77±0.07)b | (0.46±0.06)c | (0.59±0.14)c | (0.48±0.10)c | |
| CnZF-HD12 | (1.00±0.00)a | (0.92±0.05)b | (1.09±0.12)a | (0.89±0.06)b | (0.25±0.12)c | |
| CnZF-HD13 | (1.00±0.00)a | (0.64±0.04)b | (0.42±0.16)c | (0.27±0.08)c | (0.15±0.02)d | |
| CnZF-HD14 | (1.00±0.00)b | (1.12±0.05)a | (0.98±0.11)b | (0.65±0.14)c | (0.34±0.13)d | |
| CnZF-HD15 | (1.00±0.00)b | (0.77±0.13)c | (1.42±0.06)a | (1.09±0.08)b | (0.65±0.10)c | |
| CnZF-HD16 | (1.00±0.00)a | (0.60±0.15)b | (1.06±0.09)a | (0.95±0.11)a | (0.48±0.10)c | |
| CnZF-HD17 | (1.00±0.00)a | (0.84±0.11)b | (0.56±0.01)c | (0.62±0.14)c | (0.47±0.07)d | |
| CnZF-HD18 | (1.00±0.00)a | (0.88±0.06)b | (1.02±0.07)a | (0.56±0.15)c | (0.46±0.15)c | |
| CnZF-HD19 | (1.00±0.00)c | (1.77±0.09)a | (1.46±0.07)b | (0.70±0.08)d | (0.56±0.07)d | |
| [1] | 李阳阳, 李驰, 任俊洋, 等. 甘蓝型油菜苗期耐旱性综合评价与耐旱性鉴定指标筛选[J]. 中国生态农业学报: 中英文, 2021, 29(8): 1327-1338. |
| Li YY, Li C, Ren JY, et al. Comprehensive evaluation and identification trait selection of drought resistance at the seedling stage of Brassica napus L.[J]. Chin J Eco Agric, 2021, 29(8): 1327-1338. | |
| [2] |
赵红梅, 邓素芳, 杨艳君, 等. 干旱胁迫对藜麦幼苗组织解剖结构和生理特性的影响[J]. 核农学报, 2021, 35(6): 1476-1483.
doi: 10.11869/j.issn.100-8551.2021.06.1476 |
|
Zhao HM, Deng SF, Yang YJ, et al. Effect of drought stress on anatomical structure and physiological characteristics of quinoa seedlings[J]. J Nucl Agric Sci, 2021, 35(6): 1476-1483.
doi: 10.11869/j.issn.100-8551.2021.06.1476 |
|
| [3] | 王洪瑞, 敖红. 干旱胁迫对红皮云杉和嫩江云杉渗透调节及抗氧化系统的影响[J]. 东北林业大学学报, 2020, 48(8): 16-21, 32. |
| Wang HR, Ao H. Response of osmotic regulation and antioxidant system to drought stress in Korean spruce and Nenjiang spruce[J]. J Northeast For Univ, 2020, 48(8): 16-21, 32. | |
| [4] | 王爽, 赵雁. 外源ABA对干旱下4种园林地被植物生长及生理特征的影响[J]. 现代园艺, 2022, 45(1): 10-13. |
| Wang S, Zhao Y. Effects of exogenous ABA on growth and physiological characteristics of four garden ground cover plants under drought[J]. Contemp Hortic, 2022, 45(1): 10-13. | |
| [5] |
Park Y, Xu ZY, Kim SY, et al. Spatial regulation of ABCG25, an ABA exporter, is an important component of the mechanism controlling cellular ABA levels[J]. Plant Cell, 2016, 28(10): 2528-2544.
doi: 10.1105/tpc.16.00359 URL |
| [6] | 马延宏. 环境胁迫下植物细胞ABA的信号转导途径[J]. 陕西农业科学, 2009, 55(4): 120-124. |
| Ma YH. Signal transduction pathway of ABA in plant cells under environmental stress[J]. Shaanxi J Agric Sci, 2009, 55(4): 120-124. | |
| [7] | 崔喜艳, 张莹莹, 周莹. 植物响应干旱胁迫转录因子研究进展[J]. 吉林农业大学学报, 2022, 44(5): 505-511. |
| Cui XY, Zhang YY, Zhou Y. Research progress of plant transcription factors in response to drought stress[J]. J Jilin Agric Univ, 2022, 44(5): 505-511. | |
| [8] |
Muthuramalingam P, Krishnan SR, Saravanan K, et al. Genome-wide identification of major transcription factor superfamilies in rice identifies key candidates involved in abiotic stress dynamism[J]. J Plant Biochem Biotechnol, 2018, 27(3): 300-317.
doi: 10.1007/s13562-018-0440-3 |
| [9] |
Xu Y, Wang YH, Long QZ, et al. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice[J]. Planta, 2014, 239(4): 803-816.
doi: 10.1007/s00425-013-2009-7 URL |
| [10] |
Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants[J]. Physiol Plant, 2006, 126(1): 62-71.
doi: 10.1111/ppl.2006.126.issue-1 URL |
| [11] |
Tran LSP, Nakashima K, Sakuma Y, et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis[J]. Plant J, 2007, 49(1): 46-63.
doi: 10.1111/tpj.2007.49.issue-1 URL |
| [12] |
Wang H, Yin XJ, Li XQ, et al. Genome-wide identification, evolution and expression analysis of the grape(Vitis vinifera L.) zinc finger-homeodomain gene family[J]. Int J Mol Sci, 2014, 15(4): 5730-5748.
doi: 10.3390/ijms15045730 pmid: 24705465 |
| [13] |
张晋玉, 晁毛妮, 杜弘杨, 等. 大豆ZF-HD转录因子GmZHD1的克隆及表达分析[J]. 华北农学报, 2017, 32(2): 1-7.
doi: 10.7668/hbnxb.2017.02.001 |
| Zhang JY, Chao MN, Du HY, et al. Cloning and expression analysis of ZF-HD transcription factor GmZHD1 in Glycine max[J]. Acta Agric Boreali Sin, 2017, 32(2): 1-7. | |
| [14] |
Liu MY, Wang XX, Sun WJ, et al. Genome-wide investigation of the ZF-HD gene family in Tartary buckwheat(Fagopyrum tataricum)[J]. BMC Plant Biol, 2019, 19(1): 248.
doi: 10.1186/s12870-019-1834-7 |
| [15] |
宋敏, 张瑶, 王丽莹, 等. 甘蓝型油菜ZF-HD基因家族的鉴定与系统进化分析[J]. 植物学报, 2019, 54(6): 699-710.
doi: 10.11983/CBB19055 |
| Song M, Zhang Y, Wang LY, et al. Genome-wide identification and phylogenetic analysis of zinc finger homeodomain family genes in Brassica napus[J]. Chin Bull Bot, 2019, 54(6): 699-710. | |
| [16] | 胡靖康. 番茄ZF-HD转录因子的全基因组挖掘及抗逆相关基因筛选[D]. 哈尔滨: 东北农业大学, 2018. |
| Hu JK. Bioinformatic analysis and identification of some resistance-associated genes of ZF-HD gene family in tomato[D]. Harbin: Northeast Agricultural University, 2018. | |
| [17] |
Liu WP, Zhao BG, Chao Q, et al. Function analysis of ZmNAC33, a positive regulator in drought stress response in Arabidopsis[J]. Plant Physiol Biochem, 2019, 145: 174-183.
doi: 10.1016/j.plaphy.2019.10.038 URL |
| [18] | 张莉俊, 戴思兰. 菊花种质资源研究进展[J]. 植物学报, 2009, 44(5): 526-535. |
| Zhang LJ, Dai SL. Research advance on germplasm resources of Chrysanthemum × morifolium[J]. Chin Bull Bot, 2009, 44(5): 526-535. | |
| [19] |
Hu W, Ma H. Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development[J]. Plant J, 2006, 45(3): 399-422.
doi: 10.1111/tpj.2006.45.issue-3 URL |
| [20] |
Khatun K, Nath UK, Robin AHK, et al. Genome-wide analysis and expression profiling of zinc finger homeodomain(ZHD)family genes reveal likely roles in organ development and stress responses in tomato[J]. BMC Genomics, 2017, 18(1): 695.
doi: 10.1186/s12864-017-4082-y URL |
| [21] | 李朝睿. 马铃薯ZF-HD基因家族的鉴定与分析[J]. 山东农业大学学报: 自然科学版, 2021, 52(3): 384-387. |
| Li CR. Identification and analysis of ZF-HD gene family in potato[J]. J Shandong Agric Univ Nat Sci Ed, 2021, 52(3): 384-387. | |
| [22] |
Tan QKG, Irish VF. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development[J]. Plant Physiol, 2006, 140(3): 1095-1108.
doi: 10.1104/pp.105.070565 URL |
| [23] | 庞丹丹, 刘玉飞, 田易萍, 等. 茶树ZF-HD转录因子基因家族的鉴定及表达分析[J]. 南方农业学报, 2021, 52(3): 632-640. |
| Pang DD, Liu YF, Tian YP, et al. Identification and expression analysis of ZF-HD transcription factor gene family in Camellia sinensis[J]. J South Agric, 2021, 52(3): 632-640. | |
| [24] |
Liu C, Ma HZ, Zhou J, et al. TsHD1 and TsNAC1 cooperatively play roles in plant growth and abiotic stress resistance of Thellungiella halophile[J]. Plant J, 2019, 99(1): 81-97.
doi: 10.1111/tpj.2019.99.issue-1 URL |
| [25] | 马瑞芳, 陈家璐, 刘笑雨, 等. 毛竹锌指同源结构域基因家族全基因组鉴定及表达分析[J]. 农业生物技术学报, 2020, 28(4): 645-657. |
| Ma RF, Chen JL, Liu XY, et al. Genome-wide identification and expression analysis of zinc finger homologous domain gene family in Phyllostachys edulis[J]. J Agric Biotechnol, 2020, 28(4): 645-657. | |
| [26] | 庞丹丹, 刘玉飞, 田易萍, 等. 茶树ZF-HD转录因子基因家族的鉴定及表达分析[J]. 南方农业学报, 2021, 52(3): 632-640. |
| Pang DD, Liu YF, Tian YP, et al. Identification and expression analysis of ZF-HD transcription factor gene family in Camellia sinensis[J]. J South Agric, 2021, 52(3): 632-640. | |
| [27] | 孙熹微, 范海阔, 弓淑芳, 等. 椰子ZF-HD基因家族的鉴定及生物信息学分析[J]. 热带作物学报, 2020, 41(2): 284-291. |
| Sun XW, Fan HK, Gong SF, et al. Identification and bioinformatics analysis of coconut ZF-HD gene family[J]. Chin J Trop Crops, 2020, 41(2): 284-291. | |
| [28] | 刘敏. 森林草莓ZF-HD基因家族的鉴定与功能初步分析[D]. 福州: 福建农林大学, 2019. |
| Liu M. Identification and preliminary functional analysis of ZF-HD gene family in woodland strawberry[D]. Fuzhou: Fujian Agriculture and Forestry University, 2019. | |
| [29] | 李春艳. 玉米ZF-HD转录因子家族耐盐、抗旱相关基因的鉴定及特性分析[D]. 重庆: 西南大学, 2018. |
| Li CY. Identification and characterization of drought and salt resistance-related genes of ZF-HD transcription factors family in maize[D]. Chongqing: Southwest University, 2018. | |
| [30] |
Bueso E, Muñoz-Bertomeu J, Campos F, et al. ARABIDOPSIS THALIANA HOMEOBOX25 uncovers a role for gibberellins in seed longevity[J]. Plant Physiol, 2014, 164(2): 999-1010.
doi: 10.1104/pp.113.232223 URL |
| [31] |
Kim JB, Kang JY, Park MY, et al. Arabidopsis zinc finger homeodomain protein ZHD5 promotes shoot regeneration and confers other cytokinin-related phenotypes when overexpressed[J]. Plant Cell Tiss Organ Cult, 2019, 137(1): 181-185.
doi: 10.1007/s11240-018-01546-7 |
| [32] |
Liu H, Yang Y, Zhang LS. Zinc finger-homeodomain transcriptional factors(ZF-HDs)in wheat(Triticum aestivum L.): identification, evolution, expression analysis and response to abiotic stresses[J]. Plants, 2021, 10(3): 593.
doi: 10.3390/plants10030593 URL |
| [33] | Shalmani A, Muhammad I, Sharif R, et al. Zinc finger-homeodomain genes: evolution, functional differentiation, and expression profiling under flowering-related treatments and abiotic stresses in plants[J]. Evol Bioinform Online, 2019, 15: 1176934319867930. |
| [34] |
Wang WL, Wu P, Li Y, et al. Erratum to: Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage[J]. Mol Genet Genomics, 2016, 291(3): 1465.
pmid: 26965506 |
| [35] |
Wang YH, Que F, Li T, et al. DcABF3, an ABF transcription factor from carrot, alters stomatal density and reduces ABA sensitivity in transgenic Arabidopsis[J]. Plant Sci, 2021, 302: 110699.
doi: 10.1016/j.plantsci.2020.110699 URL |
| [36] |
Chen BJ, Wang Y, Hu YL, et al. Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia[J]. Plant Sci, 2005, 168(2): 493-500.
doi: 10.1016/j.plantsci.2004.09.013 URL |
| [37] |
Zhu MK, Chen GP, Zhang JL, et al. The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato(Solanum lycopersicum)[J]. Plant Cell Rep, 2014, 33(11): 1851-1863.
doi: 10.1007/s00299-014-1662-z URL |
| [38] |
Wang L, Hua DP, He JN, et al. Auxin Response Factor2(ARF2)and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis[J]. PLoS Genet, 2011, 7(7): e1002172.
doi: 10.1371/journal.pgen.1002172 URL |
| [39] |
Zhou CZ, Zhu C, Xie SY, et al. Genome-wide analysis of zinc finger motif-associated homeodomain(ZF-HD)family genes and their expression profiles under abiotic stresses and phytohormones stimuli in tea plants(Camellia sinensis)[J]. Sci Hortic, 2021, 281: 109976.
doi: 10.1016/j.scienta.2021.109976 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [3] | LIU Wen-jin, MA Rui, LIU Sheng-yan, YANG Jiang-wei, ZHANG Ning, SI Huai-jun. Cloning of StCIPK11 Gene and Analysis of Its Response to Drought Stress in Solanum tuberosum [J]. Biotechnology Bulletin, 2023, 39(9): 147-155. |
| [4] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [5] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [6] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [7] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [8] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [9] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [10] | DING Kai-xin, WANG Li-chun, TIAN Guo-kui, WANG Hai-yan, LI Feng-yun, PAN Yang, PANG Ze, SHAN Ying. Research Progress in Uniconazole Alleviating Plant Drought Damage [J]. Biotechnology Bulletin, 2023, 39(6): 1-11. |
| [11] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [12] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [13] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [14] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [15] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||