








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 1-10.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1155
HE Shan( ), WANG Zhi-kang, YANG Yang, MU Yue-wei, HUANG Yu-bi, YU Guo-wu(
), WANG Zhi-kang, YANG Yang, MU Yue-wei, HUANG Yu-bi, YU Guo-wu( )
)
Received:2020-09-13
Online:2021-05-26
Published:2021-06-11
Contact:
YU Guo-wu
E-mail:1172218473@qq.com;2002ygw@163.com
HE Shan, WANG Zhi-kang, YANG Yang, MU Yue-wei, HUANG Yu-bi, YU Guo-wu. Gene Cloning and Expression Analysis of Transcription Factor NAC78 in Maize[J]. Biotechnology Bulletin, 2021, 37(5): 1-10.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| NAC78-F | ATGGCGCCGGTCAGTTTGCCTC |
| NAC78-R | TCACTCACTGCCGCTATTTC |
| NAC78-RT-F | GGCCGTTAGGTTGGTTGAGA |
| NAC78-RT-R | TGAAGCCGCACACCTCTAAG |
| Actin-F | TCACTACGACTGCCGAGCGAG |
| Actin-R | GAGCCACCACTGAGGACAACATTAC |
Table 1 Primers used in the experiment
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| NAC78-F | ATGGCGCCGGTCAGTTTGCCTC |
| NAC78-R | TCACTCACTGCCGCTATTTC |
| NAC78-RT-F | GGCCGTTAGGTTGGTTGAGA |
| NAC78-RT-R | TGAAGCCGCACACCTCTAAG |
| Actin-F | TCACTACGACTGCCGAGCGAG |
| Actin-R | GAGCCACCACTGAGGACAACATTAC |

Fig.1 Bioinformatics analysis of NAC78 protein A: Prediction of hydrophilicity and hydrophobicity of NAC78 protein. B: NAC78 protein domain. C: Three dimensional structure prediction of NAC78 protein

Fig.2 Multiple sequence alignment of NAC78 homologous genes in different species Sorghun bicolor (SORBI_3001G529200), Setaria italica (SETIT_034541mg), Zea mays (Zm00001d027395), Oryza sativa japonica group (Os03t0119966), Triticum aestivum (Traes_CS5A02G519300), Musa acuminata (GSMUA_Achr1T02820), Populus trichocarpa (POPTR_017G139500v3), Manihot esculenta (MANES_02G081200), Gossypium raimondii (B456_007G188800), Solanum tuberosum (XP_015165583.1), Solanum lycopersicum (Solyc02g036430), Nicotiana attenuata (NW_017670850.1), Actinidia chinensis (CEY00_Acc24239), Helianthus annuus (NC_035448.1), Cucumis sativus (Csa_5G324000), Brassica napus (BnaA05g22540D), Arabidopsis thaliana (AT5G17260), Physcomitrella patens (Pp3c8_6080V3), Selaginella moellendorffii (SELMODRAFT_100973), Marchantia polymorpha (OAE31278.1). A-E: Five subdomains of NAC domain; squiggles represents α-helices, arrows represents β-sheet and TT letters represents turns
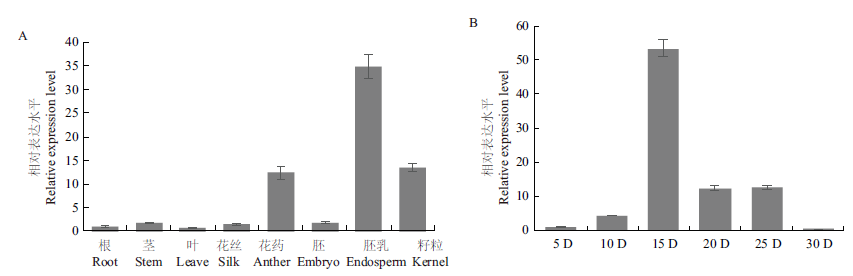
Fig.4 Relative expression of NAC78 in different tissues (A) and endosperm at different stages after pollination (B) The error line represents the standard deviation, n=3
| 元件名称Element name | 序列Sequence | 功能Function | 数量Number | 分类Classification |
|---|---|---|---|---|
| ABRE | ACGTG;TACGTG;CACGTA | 响应脱落酸 | 4 | 激素响应元件 |
| ABRE3a | TACGTG | 响应脱落酸 | 1 | |
| ABRE4 | CACGTA | 响应脱落酸 | 1 | |
| CGTCA-motif | CGTCA | 响应茉莉酸 | 3 | |
| GARE-motif | TCTGTTG | 响应赤霉素 | 1 | |
| TGA-box | TGACGTAA | 响应生长素 | 1 | |
| TGACG-motif | TGACG | 响应茉莉酸 | 3 | |
| G-Box | TACGTG | 响应光照 | 3 | 响应光照 |
| GT1-motif | TACGTG | 响应光照 | 1 | |
| Sp1 | GGGCGG | 响应光照 | 1 | |
| MBS | CAACTG | MYB转录因子结合位点 | 2 | |
| MYB | CAACCA;TAACCA;CCGTTG;TAACCA | MYB转录因子结合位点 | 7 | 转录因子结合位点 |
| O2-site | GATGACATGG | O2转录因子结合位点 | 1 | |
| CCAAT-box | CAACGG | MYBHv1转录因子结合位点 | 2 | |
| RY-element | CATGCATG | 调控种子发育 | 3 | 调控植物发育 |
| ARE | TGCTATCCG | 响应缺氧条件 | 1 | 响应非生物胁迫 |
| GC-motif | CCCCCG | 响应缺氧条件 | 2 | |
| TATA-box | TATATA | 转录起始必须 | 4 | 转录必须元件 |
| CAAT-box | CAAT;CAAAT | 转录增强元件 | 4 |
Table 2 Cis-elements in the promoter of NAC78
| 元件名称Element name | 序列Sequence | 功能Function | 数量Number | 分类Classification |
|---|---|---|---|---|
| ABRE | ACGTG;TACGTG;CACGTA | 响应脱落酸 | 4 | 激素响应元件 |
| ABRE3a | TACGTG | 响应脱落酸 | 1 | |
| ABRE4 | CACGTA | 响应脱落酸 | 1 | |
| CGTCA-motif | CGTCA | 响应茉莉酸 | 3 | |
| GARE-motif | TCTGTTG | 响应赤霉素 | 1 | |
| TGA-box | TGACGTAA | 响应生长素 | 1 | |
| TGACG-motif | TGACG | 响应茉莉酸 | 3 | |
| G-Box | TACGTG | 响应光照 | 3 | 响应光照 |
| GT1-motif | TACGTG | 响应光照 | 1 | |
| Sp1 | GGGCGG | 响应光照 | 1 | |
| MBS | CAACTG | MYB转录因子结合位点 | 2 | |
| MYB | CAACCA;TAACCA;CCGTTG;TAACCA | MYB转录因子结合位点 | 7 | 转录因子结合位点 |
| O2-site | GATGACATGG | O2转录因子结合位点 | 1 | |
| CCAAT-box | CAACGG | MYBHv1转录因子结合位点 | 2 | |
| RY-element | CATGCATG | 调控种子发育 | 3 | 调控植物发育 |
| ARE | TGCTATCCG | 响应缺氧条件 | 1 | 响应非生物胁迫 |
| GC-motif | CCCCCG | 响应缺氧条件 | 2 | |
| TATA-box | TATATA | 转录起始必须 | 4 | 转录必须元件 |
| CAAT-box | CAAT;CAAAT | 转录增强元件 | 4 |

Fig.5 Effect of ABA treatment on NAC78 expression in maize kernel The error line refers to the standard deviation. sterisk refers to significant difference (*P<0.05, **P<0.01)

Fig.6 Effects of NaCl and PEG treatment on NAC78 expression in leaves The error line represents the standard deviation; sterisk represents significant difference between 0 h. (*P<0.05, **P<0.01)
| [1] |
Souer E, Houwelinggen VA, Kloos D, et al. The noapical meris-tem gene of petunia is required for pattern formation in embryosand flowers and is expressed at meristem and primordial bound aries[J]. Cell, 1996,85(2):159-170.
pmid: 8612269 |
| [2] |
Duval M, Hsieh T, Km SY, et al. Molecular characterization of AtNAM:a member of the Arabidopsis NAC domain superfamily[J]. Plant Molecular Biology, 2002,50(2):237-248.
doi: 10.1023/A:1016028530943 URL |
| [3] |
Aida M, Ishida T, Fukaki H, et al. Genes involved in organ separation in Arabidopsis, analysis of the cup-shaped cotyledon mutant[J]. The Plant Cell, 1997,9(6):841-857.
doi: 10.1105/tpc.9.6.841 URL |
| [4] | Olsen AN, Ernst HA, Leggio LL, et al. NAC transcription factors:structurally distinct, functionally diverse[J]. Trends in Plant Science, 2005,10(2):80-87. |
| [5] |
Ooka H, Satoh K, Doi K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana[J]. DNA Research, 2003,10(6):239-247.
doi: 10.1093/dnares/10.6.239 URL |
| [6] |
Nuruzzaman M, Manimekalai R, Sharoni AM, et al. Genome-wide analysis of NAC transcription factor family in rice[J]. Gene, 2010,465(1-2):30-44.
doi: 10.1016/j.gene.2010.06.008 pmid: 20600702 |
| [7] |
Shiriga K, Sharma R, Kumar K, et al. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize[J]. Meta Gene, 2014,2:407-417.
doi: 10.1016/j.mgene.2014.05.001 URL |
| [8] |
Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis:interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes[J]. Development, 1999,126(8):1563-1570.
pmid: 10079219 |
| [9] |
Guan Q, Yue X, Zeng H, et al. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis[J]. The Plant Cell, 2014,26(1):438-453.
doi: 10.1105/tpc.113.118927 URL |
| [10] | Bu Q, Jiang H, Li CB, et al. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses[J]. Cell Reseach, 2008,18(7):756-767. |
| [11] |
Maugarny-Calès A, Gonçalves B, Jouannic S, et al. Apparition of the NAC transcription factors predates the emergence of land plants[J]. Molecular Plant, 2016,9(9):1345-1348.
doi: 10.1016/j.molp.2016.05.016 URL |
| [12] |
Wu Y, Hou J, Yu F, et al. Transcript profiling identifies NAC-domain genes involved in regulating wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana[J]. Front Plant Science, 2018,9:341.
doi: 10.3389/fpls.2018.00341 URL |
| [13] |
Lin YJ, Chen H, Li Q, et al. Reciprocal cross-regulation of VND and SND multigene TF families for wood formation in Populus trichocarpa[J]. Proc Natl Acad Sci U S A, 2017,114(45):E9722-E9729.
doi: 10.1073/pnas.1714422114 URL |
| [14] |
Xu B, Ohtani M, Yamaguchi M, et al. Contribution of NAC transcription factors to plant adaptation to land[J]. Science, 2014,343(6178):1505-1508.
doi: 10.1126/science.1248417 URL |
| [15] |
Puranik S, Sahu PP, Srivastava PS, et al. NAC proteins:regulation and role in stress tolerance[J]. Trends in Plant Science, 2012,17(6):369-381.
doi: 10.1016/j.tplants.2012.02.004 pmid: 22445067 |
| [16] |
Hu P, Zhang K, Yang C. BpNAC012 Positively regulates abiotic stress responses and secondary wall biosynjournal[J]. Plant Physiology, 2019,179(2):700-717.
doi: 10.1104/pp.18.01167 URL |
| [17] |
Hu H, Dai M, Yao J, et al. Overexpressing a NAM, ATAF, and CUC(NAC)transcription factor enhances drought resistance and salt tolerance in rice[J]. Proc Natl Acad Sci U S A, 2006,103(35):12987-12992.
doi: 10.1073/pnas.0604882103 URL |
| [18] |
He X, Zhu L, Xu L, et al. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks[J]. Plant Cell Report, 2016,35(10):2167-2179.
doi: 10.1007/s00299-016-2027-6 URL |
| [19] |
Fujita Y, Fujita M, Shinozaki K, et al. ABA-mediated transcriptional regulation in response to osmotic stress in plants[J]. Journal of Plant Research, 2011,124(4):509-525.
doi: 10.1007/s10265-011-0412-3 URL |
| [20] | Bari R, Jones J D. Role of plant hormones in plant defence responses[J]. Plant Molocular Biology, 2009,69(4):473-488. |
| [21] |
He L, Bian J, Xu J, et al. Novel maize NAC transcriptional repressor ZmNAC071 confers enhanced sensitivity to ABA and osmotic stress by downregulating stress-responsive genes in transgenic Arabidopsis[J]. Journal of Agricultural and Food Chemistry, 2019,67(32):8905-8918.
doi: 10.1021/acs.jafc.9b02331 URL |
| [22] |
Zhu Y, Yan J, Liu W, et al. Phosphorylation of a NAC transcription factor by a calcium/calmodulin-dependent protein kinase regulates abscisic acid-induced antioxidant defense in maize[J]. Plant Physiology, 2016,171(3):1651-1664.
doi: 10.1104/pp.16.00168 URL |
| [23] |
Shen J, Lv B, Luo L, et al. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice[J]. Science Report, 2017,7:40641.
doi: 10.1038/srep40641 URL |
| [24] | 杨欢. 灌浆期高温干旱胁迫影响糯玉米籽粒产量形成的生理机制[D]. 扬州:扬州大学, 2017. |
| Yang H. Physiological of high temperature and drought stress during grain filling on yield formation in waxy maize[D]. Yangzhou:Yangzhou University, 2017. | |
| [25] | 李叶蓓, 陶洪斌, 王若男, 等. 干旱对玉米穗发育及产量的影响[J]. 中国生态农业学报, 2015,23(4):383-391. |
| Li YB, Tao HB, Wang RN, et al. Effect of drought on ear development and yield of maize[J]. Chinese Journal of Eco-Agriculture, 2015,23(4):383-391. | |
| [26] | 匡朴. 盐胁迫对不同耐盐性玉米品种萌发、苗期生长及产量的影响[D]. 泰安:山东农业大学, 2018. |
| Kuang P. Effects of salt stress on seed germination, seeding growth and yield of maize variaties with different salt tolerance[D]. Tai’an:Shandong Agricultural University, 2018. | |
| [27] |
Zhang L, Li XH, Gao Z, et al. Regulation of maize kernel weight and carbohydrate metabolism by abscisic acid applied at the early and middle post-pollination stages in vitro[J]. Journal of Plant Physiology, 2017,216:1-10.
doi: S0176-1617(17)30125-6 pmid: 28544894 |
| [28] |
Chen J, Zeng B, Zhang M, et al. Dynamic transcriptome landscape of maize embryo and endosperm development[J]. Plant Physiology, 2014,166(1):252-264.
doi: 10.1104/pp.114.240689 URL |
| [29] |
Ernst HA, Olsen AN, Larsen S, et al. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors[J]. EMBO Report, 2004,5(3):297-303.
doi: 10.1038/sj.embor.7400093 URL |
| [30] | Mathew IE, Agarwal P. May the fittest protein evolve:favoring the plant-specific origin and expansion of NAC transcription factors[J]. Bioessays, 2018,40(8):e1800018. |
| [31] |
Kang M, Kim S, Kim HJ, et al. The C-domain of the NAC transcription factor ANAC019 is necessary for pH-tuned DNA binding through a histidine switch in the N-domain[J]. Cell Report, 2018,22(5):1141-1150.
doi: 10.1016/j.celrep.2018.01.002 URL |
| [32] |
Pereira-Santana A, Alcaraz LD, Castaño E, et al. Comparative Genomics of NAC transcriptional factors in angiosperms:implications for the adaptation and diversification of flowering plants[J]. PLoS One, 2015,10(11):e0141866.
doi: 10.1371/journal.pone.0141866 URL |
| [33] |
Furuta KM, Yadav SR, Lehesranta S, et al. Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation[J]. Science, 2014,345(6199):933-937.
doi: 10.1126/science.1253736 URL |
| [34] | Eleftheriou EP. Ultrastructural studies on protophloem sieve elements in Triticum aestivum L. nuclear degeneration[J]. Journal of Ultrastructure Research & Molecular Structure Research, 1986,95(1):47-60. |
| [35] |
Groover A. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synjournal[J]. Plant Physiology, 1999,119(2):375-384.
pmid: 9952432 |
| [36] |
Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals[J]. Plant Molecular Biology, 1999,39(5):915-926.
doi: 10.1023/A:1006134027834 URL |
| [37] | 李睿. 稻麦淀粉胚乳细胞程序性死亡和淀粉质体的发生发育[D]. 武汉:华中农业大学, 2004. |
| Li R. The programmed cell death of starchy endosperm and the genesis and development of amyloplast in rice and wheat[D]. Wuhan:Huazhong. Agricultural University, 2004. | |
| [38] |
Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants[J]. Current Opinion in Plant Biology, 2014,21:133-139.
doi: S1369-5266(14)00106-X pmid: 25104049 |
| [39] | Shao HB, Liang ZS, Shao MG. Roles of ABA signal transduction during higher plant seed development and germination[J]. Forest Ecosystems, 2003,5(4):45-53. |
| [40] |
Rook F, Corke F, Card R, et al. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signaling[J]. Plant Journal, 2001,26(4):421-433.
pmid: 11439129 |
| [41] | Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and indepedent abiotic stress signaling[J]. Biologia Plantarum(Prague), 2010,54(2):201-212. |
| [42] |
Bossi F, Cordoba E, Patricia D, et al. The Arabidopsis ABA-INSENSITIVE(ABI)4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2. 2 genes during sugar signaling[J]. The Plant Journal, 2009,59(3):359-374.
doi: 10.1111/tpj.2009.59.issue-3 URL |
| [43] |
Ramon M, Rolland F, Thevelein JM, et al. ABI4 mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown[J]. Plant Molecular Biology, 2007,63(2):195-206.
doi: 10.1007/s11103-006-9082-2 URL |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [5] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [8] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [9] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [10] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [11] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [12] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [13] | SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato [J]. Biotechnology Bulletin, 2023, 39(5): 233-242. |
| [14] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [15] | WANG Hai-long, LI Yu-qian, WANG Bo, XING Guo-fang, ZHANG Jie-wei. Isolation and Expression Analysis of SiMAPK3 in Setaria italica L. [J]. Biotechnology Bulletin, 2023, 39(3): 123-132. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||