








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 233-242.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1241
Previous Articles Next Articles
SHI Jian-lei1,2( ), ZAI Wen-shan1, SU Shi-wen1, FU Cun-nian1, XIONG Zi-li1(
), ZAI Wen-shan1, SU Shi-wen1, FU Cun-nian1, XIONG Zi-li1( )
)
Received:2022-10-09
Online:2023-05-26
Published:2023-06-08
Contact:
XIONG Zi-li
E-mail:sjlhebau@163.com;273493129@qq.com
SHI Jian-lei, ZAI Wen-shan, SU Shi-wen, FU Cun-nian, XIONG Zi-li. Identification and Expression Analysis of miRNA Related to Bacterial Wilt Resistance in Tomato[J]. Biotechnology Bulletin, 2023, 39(5): 233-242.
| 基因 Gene | 序列 Sequence(5'-3') | 引物 Primer(5'-3') |
|---|---|---|
| SlU6 | GTCCCTTCGGGGACATCCGATAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCACAAATCGAGAAATGGTCCAAAATTTT | F: GGGACATCCGATAAAATTGG R: TTGGACCATTTCTCGATTTGT |
| novel_miR_13 | GGCGGAUGUAGCCAAGUGGA | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCACTT F: ACACTCCAGCTGGGGGCGGATGTAGCC |
| sly-miR172c | AGAATCTTGATGATGCTGCAG | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTGCAGC F: ACACTCCAGCTGGGAGAATCTTGATGAT |
| sly-miR172d | GGAATCTTGATGATGCTGCAG | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTGCAGC F: ACACTCCAGCTGGGGGAATCTTGATGAT |
| sly-miR396a-5p | TTCCACAGCTTTCTTGAACTG | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGTTCAA F: ACACTCCAGCTGGGTTCCACAGCTTTC |
| All R: TGGTGTCGTGGAGTCG |
Table 1 Primers used for RT-qPCR
| 基因 Gene | 序列 Sequence(5'-3') | 引物 Primer(5'-3') |
|---|---|---|
| SlU6 | GTCCCTTCGGGGACATCCGATAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCACAAATCGAGAAATGGTCCAAAATTTT | F: GGGACATCCGATAAAATTGG R: TTGGACCATTTCTCGATTTGT |
| novel_miR_13 | GGCGGAUGUAGCCAAGUGGA | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCACTT F: ACACTCCAGCTGGGGGCGGATGTAGCC |
| sly-miR172c | AGAATCTTGATGATGCTGCAG | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTGCAGC F: ACACTCCAGCTGGGAGAATCTTGATGAT |
| sly-miR172d | GGAATCTTGATGATGCTGCAG | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTGCAGC F: ACACTCCAGCTGGGGGAATCTTGATGAT |
| sly-miR396a-5p | TTCCACAGCTTTCTTGAACTG | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGTTCAA F: ACACTCCAGCTGGGTTCCACAGCTTTC |
| All R: TGGTGTCGTGGAGTCG |
| 样本Library | 净读数Clean reads/M | 净碱基数Clean bases/Gb | GC含量GC content/% | Q30/% | 无注释Unannotated | 比对读数Mapped reads |
|---|---|---|---|---|---|---|
| RC02 | 14.31 | 0.32 | 41.86 | 95.26 | 10.81 | 7.08 |
| RC14 | 16.69 | 0.38 | 42.34 | 95.26 | 11.86 | 7.68 |
| RT06 | 12.93 | 0.30 | 42.68 | 95.61 | 8.85 | 5.72 |
| RT18 | 10.84 | 0.24 | 41.95 | 95.57 | 7.92 | 5.11 |
| SC04 | 14.57 | 0.33 | 42.10 | 95.68 | 10.22 | 7.31 |
| SC16 | 16.54 | 0.37 | 43.15 | 95.75 | 10.79 | 7.77 |
| ST08 | 11.32 | 0.26 | 42.76 | 95.55 | 7.68 | 5.14 |
| ST20 | 15.56 | 0.35 | 41.62 | 95.77 | 11.52 | 7.75 |
Table 2 Statistics of sRNA sequencing reads in the eight tomato libraries
| 样本Library | 净读数Clean reads/M | 净碱基数Clean bases/Gb | GC含量GC content/% | Q30/% | 无注释Unannotated | 比对读数Mapped reads |
|---|---|---|---|---|---|---|
| RC02 | 14.31 | 0.32 | 41.86 | 95.26 | 10.81 | 7.08 |
| RC14 | 16.69 | 0.38 | 42.34 | 95.26 | 11.86 | 7.68 |
| RT06 | 12.93 | 0.30 | 42.68 | 95.61 | 8.85 | 5.72 |
| RT18 | 10.84 | 0.24 | 41.95 | 95.57 | 7.92 | 5.11 |
| SC04 | 14.57 | 0.33 | 42.10 | 95.68 | 10.22 | 7.31 |
| SC16 | 16.54 | 0.37 | 43.15 | 95.75 | 10.79 | 7.77 |
| ST08 | 11.32 | 0.26 | 42.76 | 95.55 | 7.68 | 5.14 |
| ST20 | 15.56 | 0.35 | 41.62 | 95.77 | 11.52 | 7.75 |
| 样本 Library | 已知miRNA Known miRNA | 新miRNA Novel miRNA | 总数 Total | 新miRNA百分率 Novel miRNA percentage/% |
|---|---|---|---|---|
| RC02 | 134 | 193 | 327 | 59.02 |
| RC14 | 138 | 193 | 331 | 58.31 |
| RT06 | 136 | 192 | 328 | 58.54 |
| RT18 | 132 | 190 | 322 | 59.01 |
| SC04 | 137 | 193 | 330 | 58.48 |
| SC16 | 136 | 193 | 329 | 58.66 |
| ST08 | 134 | 193 | 327 | 59.02 |
| ST20 | 136 | 193 | 329 | 58.66 |
| Total | 143 | 193 | 336 | 57.44 |
Table 3 Statistics of known and new miRNA in each lib-rary
| 样本 Library | 已知miRNA Known miRNA | 新miRNA Novel miRNA | 总数 Total | 新miRNA百分率 Novel miRNA percentage/% |
|---|---|---|---|---|
| RC02 | 134 | 193 | 327 | 59.02 |
| RC14 | 138 | 193 | 331 | 58.31 |
| RT06 | 136 | 192 | 328 | 58.54 |
| RT18 | 132 | 190 | 322 | 59.01 |
| SC04 | 137 | 193 | 330 | 58.48 |
| SC16 | 136 | 193 | 329 | 58.66 |
| ST08 | 134 | 193 | 327 | 59.02 |
| ST20 | 136 | 193 | 329 | 58.66 |
| Total | 143 | 193 | 336 | 57.44 |
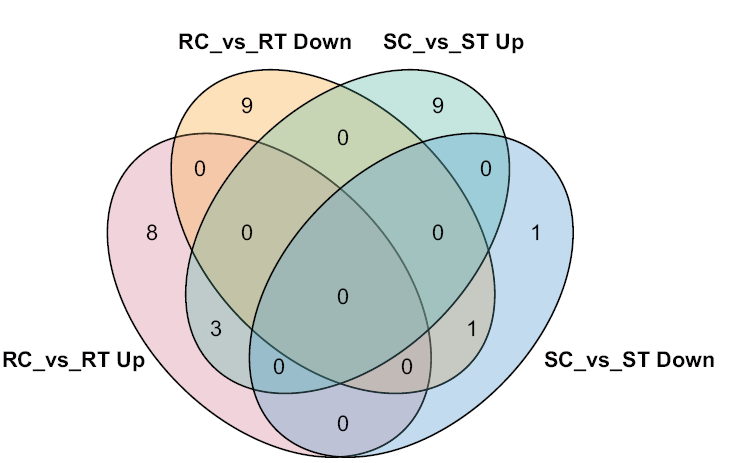
Fig. 1 Differentially expressed miRNA in resistance and susceptible tomato lines Up and Down indicate up-regulated and down-regulated expression, respectively
| 类型 Type | 所有miRNA All miRNA | 具有靶基因的miRNA miRNA with targets | 靶基因 Target genes |
|---|---|---|---|
| Known miRNA | 143 | 134 | 2 268 |
| Novel miRNA | 193 | 159 | 1 943 |
| 总数 Total | 336 | 293 | 3 960 |
Table 4 Statistics of miRNA targets
| 类型 Type | 所有miRNA All miRNA | 具有靶基因的miRNA miRNA with targets | 靶基因 Target genes |
|---|---|---|---|
| Known miRNA | 143 | 134 | 2 268 |
| Novel miRNA | 193 | 159 | 1 943 |
| 总数 Total | 336 | 293 | 3 960 |
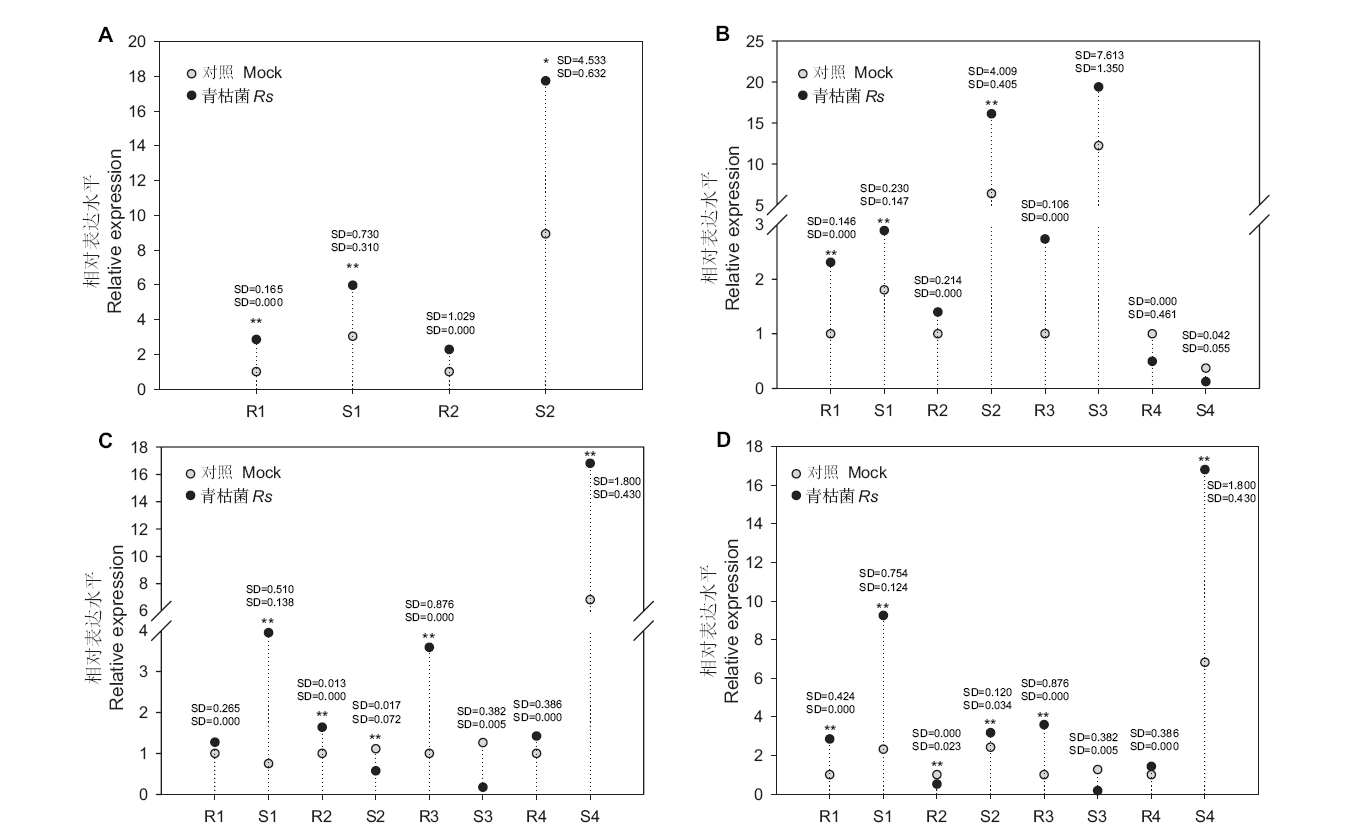
Fig. 5 Relative expressions of ten selected miRNA-targets in resistant and susceptible tomato lines with R. solanacearum infection R and S represent resistant and susceptible tomato lines, respectively. * and ** indicate statistically significant differences (P < 0.05 and P < 0.01). SD stands for standard deviation. A: 1: novel_miR_13; 2: Solyc09g091370.4. B:1: sly-miR396a-5p; 2: Solyc02g032870.4; 3: Solyc07g041640.3; 4: Solyc09g009200.3. C: 1: sly-miR172c; 2: Solyc04g064620.4; 3: Solyc10g006710.4; 4: Solyc12g099450.3. D: 1: sly-miR172d; 2: Solyc09g075550.3; 3: Solyc10g006710.4; 4: Solyc12g099450.3
| [1] |
Budak H, Zhang B. microRNAs in model and complex organisms[J]. Funct Integr Genomics, 2017, 17(2/3): 121-124.
doi: 10.1007/s10142-017-0544-1 URL |
| [2] |
张翠桔, 莫蓓莘, 陈雪梅, 等. 植物miRNA作用方式的分子机制研究进展[J]. 生物技术通报, 2020, 36(7): 1-14.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0262 |
| Zhang CJ, Mo BX, Chen XM, et al. Advances on the molecular action mechanisms of plant miRNA[J]. Biotechnol Bull, 2020, 36(7): 1-14. | |
| [3] |
Zhang Y, Xia R, Kuang HH, et al. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them[J]. Mol Biol Evol, 2016, 33(10): 2692-2705.
doi: 10.1093/molbev/msw154 pmid: 27512116 |
| [4] |
Li Y, Zhang QQ, Zhang JG, et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity[J]. Plant Physiol, 2010, 152(4): 2222-2231.
doi: 10.1104/pp.109.151803 pmid: 20164210 |
| [5] |
Wang YX, Wang Q, Gao LP, et al. Parsing the regulatory network between small RNAs and target genes in ethylene pathway in tomato[J]. Front Plant Sci, 2017, 8: 527.
doi: 10.3389/fpls.2017.00527 pmid: 28443119 |
| [6] |
李琳琳, 金华, 刘斯超, 等. 番茄茉莉酸缺失突变体灰霉菌侵染响应miRNA及其表达分析[J]. 园艺学报, 2020, 47(7): 1323-1334.
doi: 10.16420/j.issn.0513-353x.2020-0162 |
| Li LL, Jin H, Liu SC, et al. Expressied analysis of miRNA with tomato JA deficient mutant reponse to Botrytis cinerea infection[J]. Acta Hortic Sin, 2020, 47(7): 1323-1334. | |
| [7] |
Bazzini AA, Hopp HE, Beachy RN, et al. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development[J]. Proc Natl Acad Sci USA, 2007, 104(29): 12157-12162.
doi: 10.1073/pnas.0705114104 pmid: 17615233 |
| [8] |
Li Y, Cao XL, Zhu Y, et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases[J]. New Phytol, 2019, 222(3): 1507-1522.
doi: 10.1111/nph.2019.222.issue-3 URL |
| [9] |
Yang L, Huang H. Roles of small RNAs in plant disease resistance[J]. J Integr Plant Biol, 2014, 56(10): 962-970.
doi: 10.1111/jipb.12200 |
| [10] |
Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, et al. Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection[J]. Front Plant Sci, 2017, 8: 378.
doi: 10.3389/fpls.2017.00378 pmid: 28382044 |
| [11] |
Feng JL, Wang YW, Lin RH, et al. Altered expression of microRNAs and target mRNAs in tomato root and stem tissues upon different viral infection[J]. J Phytopathol, 2013, 161(2): 107-119.
doi: 10.1111/jph.12030 URL |
| [12] |
Xu XW, Zhong CL, Tan M, et al. Identification of microRNAs and their targets that respond to powdery mildew infection in cucumber by small RNA and degradome sequencing[J]. Front Genet, 2020, 11: 246.
doi: 10.3389/fgene.2020.00246 pmid: 32273882 |
| [13] | Shi JL, Zai WS, Xiong ZL, et al. Small RNA profiling reveals a role of miRNAs in response to Ralstonia solanacearum infection in tomato[J]. J Plant Growth Regul, 2022: 1-14. |
| [14] |
Friedländer MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades[J]. Nucleic Acids Res, 2012, 40(1): 37-52.
doi: 10.1093/nar/gkr688 pmid: 21911355 |
| [15] |
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550.
doi: 10.1186/s13059-014-0550-8 URL |
| [16] |
Allen E, Xie ZX, Gustafson AM, et al. microRNA-directed phasing during trans-acting siRNA biogenesis in plants[J]. Cell, 2005, 121(2): 207-221.
doi: 10.1016/j.cell.2005.04.004 pmid: 15851028 |
| [17] |
Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology[J]. Nat Genet, 2000, 25(1): 25-29.
doi: 10.1038/75556 pmid: 10802651 |
| [18] | Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome[J]. Nucleic Acids Res, 2004, 32(Data-base issue): D277-D280. |
| [19] |
Garg V, Khan AW, Kudapa H, et al. Integrated transcriptome, small RNA and degradome sequencing approaches provide insights into Ascochyta blight resistance in chickpea[J]. Plant Biotechnol J, 2019, 17(5): 914-931.
doi: 10.1111/pbi.2019.17.issue-5 URL |
| [20] |
Sunkar R, Zhou XF, Zheng Y, et al. Identification of novel and candidate miRNAs in rice by high throughput sequencing[J]. BMC Plant Biol, 2008, 8: 25.
doi: 10.1186/1471-2229-8-25 pmid: 18312648 |
| [21] |
Kohli D, Joshi G, Deokar AA, et al. Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing[J]. PLoS One, 2014, 9(10): e108851.
doi: 10.1371/journal.pone.0108851 URL |
| [22] |
Hua CL, Zhao JH, Guo HS. Trans-kingdom RNA silencing in plant-fungal pathogen interactions[J]. Mol Plant, 2018, 11(2): 235-244.
doi: S1674-2052(17)30370-2 pmid: 29229568 |
| [23] |
Li F, Pignatta D, Bendix C, et al. microRNA regulation of plant innate immune receptors[J]. PNAS, 2012, 109(5): 1790-1795.
doi: 10.1073/pnas.1118282109 pmid: 22307647 |
| [24] |
Shivaprasad PV, Chen HM, Patel K, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs[J]. Plant Cell, 2012, 24(3): 859-874.
doi: 10.1105/tpc.111.095380 URL |
| [25] |
Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis[J]. Planta, 2009, 229(4): 1009-1014.
doi: 10.1007/s00425-009-0889-3 pmid: 19148671 |
| [26] |
Jin WB, Wu FL, Xiao L, et al. Microarray-based analysis of tomato miRNA regulated by Botrytis cinerea[J]. J Plant Growth Regul, 2012, 31(1): 38-46.
doi: 10.1007/s00344-011-9217-9 URL |
| [27] |
Jin WB, Wu FL. Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves[J]. BMC Plant Biol, 2015, 15: 1.
doi: 10.1186/s12870-014-0410-4 URL |
| [28] |
Luan YS, Cui J, Li J, et al. Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum[J]. Planta, 2018, 247(1): 127-138.
doi: 10.1007/s00425-017-2773-x URL |
| [29] |
Chen L, Meng J, Zhai JM, et al. microRNA396a-5p and-3p induce tomato disease susceptibility by suppressing target genes and upregulating salicylic acid[J]. Plant Sci, 2017, 265: 177-187.
doi: S0168-9452(17)30400-4 pmid: 29223339 |
| [30] |
Ji HM, Zhao M, Gao Y, et al. FRG3, a target of slmiR482e-3p, provides resistance against the fungal pathogen Fusarium oxysporum in tomato[J]. Front Plant Sci, 2018, 9: 26.
doi: 10.3389/fpls.2018.00026 URL |
| [31] |
Hong YH, Meng J, He XL, et al. Overexpression of miR482c in tomato induces enhanced susceptibility to late blight[J]. Cells, 2019, 8(8): 822.
doi: 10.3390/cells8080822 URL |
| [32] |
Li Y, Lu YG, Shi Y, et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae[J]. Plant Physiol, 2014, 164(2): 1077-1092.
doi: 10.1104/pp.113.230052 URL |
| [33] |
Zhang C, Ding ZM, Wu KC, et al. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice[J]. Mol Plant, 2016, 9(9): 1302-1314.
doi: S1674-2052(16)30127-7 pmid: 27381440 |
| [34] |
Su YC, Zhang YY, Huang N, et al. Small RNA sequencing reveals a role for sugarcane miRNAs and their targets in response to Sporisorium scitamineum infection[J]. BMC Genomics, 2017, 18(1): 325.
doi: 10.1186/s12864-017-3716-4 URL |
| [1] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [2] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [3] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [4] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| [5] | LV Yu-jing, WU Dan-dan, KONG Chun-yan, YANG Yu, GONG Ming. Genome-wide Identification of XTH Gene Family and Their Interacting miRNAs and Possible Roles in Low Temperature Adaptation in Jatropha curcas L. [J]. Biotechnology Bulletin, 2023, 39(2): 147-160. |
| [6] | CHEN Hao-ting, ZHANG Yu-jing, LIU Jie, DAI Ze-min, LIU Wei, SHI Yu, ZHANG Yi, LI Tian-lai. Functional Analysis of WRKY6 Gene in Tomato Under Low-phosphorus Stress [J]. Biotechnology Bulletin, 2023, 39(10): 136-147. |
| [7] | YIN Guo-ying, LIU Chang, CHANG Yong-chun, YU Wang-jie, WANG Bing, ZHANG Pan, GUO Yu-shuang. Identification of the Cysteine Protease Family and Corresponding miRNAs in Nicotiana tabacum L. and Their Responses to PVY [J]. Biotechnology Bulletin, 2023, 39(10): 184-196. |
| [8] | WANG Nan-nan, WANG Wen-jia, ZHU Qiang. Research Progress of microRNAs in Plant Stress Responses [J]. Biotechnology Bulletin, 2022, 38(8): 1-11. |
| [9] | MA Xin-xin, XU Yang, ZHAO Huan-huan, HUO Zhao-yan, WANG Shu-bin, ZHONG Feng-lin. Identification of Tomato 4CL Gene Family and Expression Analysis Under Nitrogen Treatment [J]. Biotechnology Bulletin, 2022, 38(4): 163-173. |
| [10] | ZHANG Hong-yan, LIN Guo-li, LI Ru-lian, JI Xiao-qi. Screening of Antagonist Against Tomato Fruit Rot and Their Preservation Qualities on Tomato [J]. Biotechnology Bulletin, 2022, 38(3): 69-78. |
| [11] | LIU Chao, CHU Hong-long, WU Li-fang, TANG Li-zhou, HAN Li-hong. Regulation Mechanism of Phosphate Homeostasis in Plants [J]. Biotechnology Bulletin, 2022, 38(2): 184-194. |
| [12] | LI Xiao-fan, GENG Dan-dan, BI Yu-lin, JIANG Yong, WANG Zhi-xiu, CHANG Guo-bin, CHEN Guo-hong, BAI Hao. Research Progress in Unconventional miRNA Functions [J]. Biotechnology Bulletin, 2022, 38(12): 1-10. |
| [13] | LI Ting-ting, DENG Xu-hui, LI Ruo-chen, LIU Hong-jun, SHEN Zong-zhuan, LI Rong, SHEN Qi-rong. Effects of Ralstonia solanacearum Infection on Soil Fungal Community Diversity [J]. Biotechnology Bulletin, 2022, 38(10): 195-203. |
| [14] | HONG Jun, WEI Xia-yi, JI Bing-jie, YE Yan-xin, CHENG Tian-ci. Change of Differentially Expressed Genes and SNP Before or After Pseudomonas aeruginosa Resistance to Tachyplesin I [J]. Biotechnology Bulletin, 2021, 37(9): 191-202. |
| [15] | CHEN Li-jie, YANG Fan, FAN Hai-yan, ZHAO Di, WANG Yuan-yuan, ZHU Xiao-feng, LIU Xiao-yu, DUAN Yu-xi. Advances of Non-coding RNA in Interactions Among Biocontrol Bacteria and Plant Nematodes and Host [J]. Biotechnology Bulletin, 2021, 37(7): 65-70. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||