








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (7): 107-117.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0599
Previous Articles Next Articles
GAO Bo( ), MA Juan, LI Xiu-hua, LI Jiao-sheng, WANG Rong-yan, CHEN Shu-long(
), MA Juan, LI Xiu-hua, LI Jiao-sheng, WANG Rong-yan, CHEN Shu-long( )
)
Received:2021-05-08
Online:2021-07-26
Published:2021-08-13
Contact:
CHEN Shu-long
E-mail:gaobo89@163.com;chenshulong65@163.com
GAO Bo, MA Juan, LI Xiu-hua, LI Jiao-sheng, WANG Rong-yan, CHEN Shu-long. Cloning and Functional Analysis of Dd-mel-26 Gene from Ditylenchus destructor[J]. Biotechnology Bulletin, 2021, 37(7): 107-117.
| 引物Primer | 序列信息Sequence(5'-3') | 备注Remarks |
|---|---|---|
| mel-26F | GATGCCTTCTGAATCTGTTG | cDNA中间区段扩增 |
| mel-26R | GCAGTAAGAACCAGGAACAG | |
| mel26-5GSP1 | GTCGGAATGTGACGGGTTTG | 5'RACE引物 |
| mel26-5GSP2 | TTTCCGTTTGTGAGGCGTTG | |
| mel26-3GSP1 | ATCGGGCAGATTTGTTACGC | 3'RACE引物 |
| mel26-3GSP2 | CCCGCAGAGCTTTCTTCGTC | |
| QT | CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT | RACE加帽引物 |
| QO | CCAGTGAGCAGAGTGACG | |
| QI | GAGGACTCGAGCTCAAGC | |
| mel-26RT | CACAAGAACGGTGATGCTAT | 5'RACE反转录引物 |
| mel-26GF | GATGCCTTCTGAATCTGTTG | 基因组序列扩增 |
| mel-26GR | ACATCACAGACATAGCCAGA | |
| 26T7aF | TAATACGACTCACTATAGGGTGCCTTCTGAATCTGTTGCC | dsRNA合成模板扩增 |
| 26T7aR | TAATACGACTCACTATAGGGGAATGTGACGGGTTTGGATG | |
| 26T7bF | TAATACGACTCACTATAGGGTCCAAACCCGTCACATTCCG | |
| 26T7bR | TAATACGACTCACTATAGGGACGAGCTGCGTCCGCAATAA | |
| 26T7cF | TAATACGACTCACTATAGGGCGTCCGAAATCAACGAACTT | |
| 26T7cR | TAATACGACTCACTATAGGGCGGAGATGGAACTTAGAGGG | |
| GFP_L1 | TAATACGACTCACTATAGGGGACTTCTTCAAGTCCGCCAT | 阴性对照dsRNA模板合成引物 |
| GFP_R1 | TAATACGACTCACTATAGGGAAGTTCACCTTGATGCCGTT | |
| mel26q_L | TTGAAATGCGAATTCATCCA | qPCR扩增引物 |
| mel26q_R | TCATAATTGCAAACGCCAAG | |
| actin_L | AGGACTTGTACGCCAACACC | |
| actin_R | ACATTTGCTGGAAGGTGGAG | |
| mel26-probe-F | TCCAAACCCGTCACATTCCG | Southern blotting探针引物 |
| mel26-probe-R | ACGAGCTGCGTCCGCAATAA |
Table 1 Primer information used in this study
| 引物Primer | 序列信息Sequence(5'-3') | 备注Remarks |
|---|---|---|
| mel-26F | GATGCCTTCTGAATCTGTTG | cDNA中间区段扩增 |
| mel-26R | GCAGTAAGAACCAGGAACAG | |
| mel26-5GSP1 | GTCGGAATGTGACGGGTTTG | 5'RACE引物 |
| mel26-5GSP2 | TTTCCGTTTGTGAGGCGTTG | |
| mel26-3GSP1 | ATCGGGCAGATTTGTTACGC | 3'RACE引物 |
| mel26-3GSP2 | CCCGCAGAGCTTTCTTCGTC | |
| QT | CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT | RACE加帽引物 |
| QO | CCAGTGAGCAGAGTGACG | |
| QI | GAGGACTCGAGCTCAAGC | |
| mel-26RT | CACAAGAACGGTGATGCTAT | 5'RACE反转录引物 |
| mel-26GF | GATGCCTTCTGAATCTGTTG | 基因组序列扩增 |
| mel-26GR | ACATCACAGACATAGCCAGA | |
| 26T7aF | TAATACGACTCACTATAGGGTGCCTTCTGAATCTGTTGCC | dsRNA合成模板扩增 |
| 26T7aR | TAATACGACTCACTATAGGGGAATGTGACGGGTTTGGATG | |
| 26T7bF | TAATACGACTCACTATAGGGTCCAAACCCGTCACATTCCG | |
| 26T7bR | TAATACGACTCACTATAGGGACGAGCTGCGTCCGCAATAA | |
| 26T7cF | TAATACGACTCACTATAGGGCGTCCGAAATCAACGAACTT | |
| 26T7cR | TAATACGACTCACTATAGGGCGGAGATGGAACTTAGAGGG | |
| GFP_L1 | TAATACGACTCACTATAGGGGACTTCTTCAAGTCCGCCAT | 阴性对照dsRNA模板合成引物 |
| GFP_R1 | TAATACGACTCACTATAGGGAAGTTCACCTTGATGCCGTT | |
| mel26q_L | TTGAAATGCGAATTCATCCA | qPCR扩增引物 |
| mel26q_R | TCATAATTGCAAACGCCAAG | |
| actin_L | AGGACTTGTACGCCAACACC | |
| actin_R | ACATTTGCTGGAAGGTGGAG | |
| mel26-probe-F | TCCAAACCCGTCACATTCCG | Southern blotting探针引物 |
| mel26-probe-R | ACGAGCTGCGTCCGCAATAA |
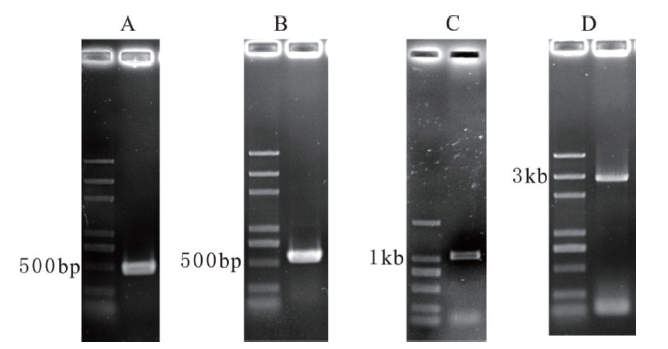
Fig. 1 Amplification results of the cDNA and genomic sequence of Dd-mel-26 A:3' RACE;B:5' RACE;C:intermediate region of the cDNA;D:Dd-mel-26 genomic sequence
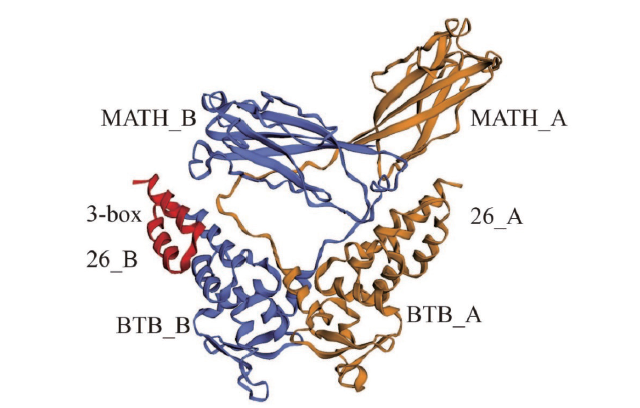
Fig. 4 Tertiary structure of Dd-MEL-26 protein The orange monomer is 26_ A,the violet monomer is 26_ B,the two red α-helix structures are 3-box domains,and the butterfly-like structure rich in β-strand in the upper part of the structure is the MATH domain(MATH_ A and MATH_B),and the butterfly-like structure rich in α-helix in the lower part is BTB domain(BTB_ A and BTB_B)

Fig. 5 Neighbor-Joining tree calculated from the sequences of Dd-mel-26 amino acid sequence Only bootstrap values more than 70% are listed at the branches points
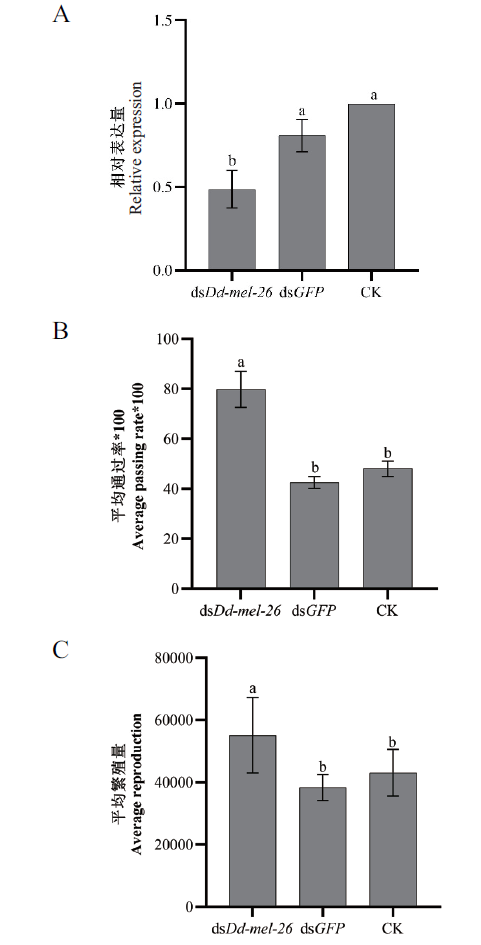
Fig. 9 Relative expressions of target gene(A),average passing rate in sand column(B),and average reproduction(C)in the Dd-mel-26 dsRNA-treated nematodes
| [1] | 丁再福, 林茂松. 甘薯、马铃薯和薄荷上的茎线虫的鉴定[J]. 植物保护学报, 1982, 9(3):169-172, 219. |
| Ding ZF, Lin MS. Identification of the stem nematodes from the sweet-potatoes, potatoes, and mints in China[J]. J Plant Prot, 1982, 9(3):169-172, 219. | |
| [2] | 刘维志, 刘清利, 尼秀媚. 马铃薯茎线虫Ditylenchus destructor Thorne, 1945的描述[J]. 莱阳农学院学报, 2003, 20(1):1-3. |
| Liu WZ, Liu QL, Ni XM. Description of stem nematodes:Ditylenchus destructor thorne, 1945[J]. J Laiyang Agric Coll, 2003, 20(1):1-3. | |
| [3] | 刘先宝, 葛建军, 谭志琼, 等. 马铃薯腐烂茎线虫在国内危害马铃薯的首次报道[J]. 植物保护, 2006, 32(6):157-158. |
| Liu XB, Ge JJ, Tan ZQ, et al. First report of Ditylenchus destructor infesting potato in China[J]. Plant Prot, 2006, 32(6):157-158. | |
| [4] | 周忠, 马代夫. 甘薯茎线虫病的研究现状和展望[J]. 杂粮作物, 2003, 23(5):288-290. |
| Zhou Z, Ma DF. Research status and prospect of sweet potato stem nematode disease[J]. Rain Fed Crops, 2003, 23(5):288-290. | |
| [5] | 王欣, 李强, 曹清河, 等. 中国甘薯产业和种业发展现状与未来展望[J]. 中国农业科学, 2021, 54(3):483-492. |
| Wang X, Li Q, Cao QH, et al. Current status and future prospective of sweetpotato production and seed industry in China[J]. Sci Agric Sin, 2021, 54(3):483-492. | |
| [6] | 王治文, 高翔, 马德君, 等. 核酸农药——极具潜力的新型植物保护产品[J]. 农药学学报, 2019, 21(Z1):681-691. |
| Wang ZW, Gao X, Ma DJ, et al. Nucleic acid pesticides—the new plant protection products with great potential[J]. Chin J Pestic Sci, 2019, 21(Z1):681-691. | |
| [7] | 李宇, 谢辉, 徐春玲, 等. Cathepsin B基因的沉默对香蕉穿孔线虫繁殖力的影响[J]. 中国农业科学, 2010, 43(8):1608-1616. |
| Li Y, Xie H, Xu CL, et al. RNAi effect of Cathepsin B gene on reproduction of Radopholus similis[J]. Sci Agric Sin, 2010, 43(8):1608-1616. | |
| [8] | 彭焕. 马铃薯腐烂茎线虫内切葡聚糖酶新基因的克隆及基于RNAi的功能初步研究[D]. 北京:中国农业科学院, 2009. |
| Peng H. Cloning two new genes of β-1, 4-endoglucanase from Ditylenchus destructor and functional analysis with RNA interference[D]. Beijing:Chinese Academy of Agricultural Sciences, 2009. | |
| [9] |
Dalzell JJ, Warnock ND, Stevenson MA, et al. Short interfering RNA-mediated knockdown of drosha and pasha in undifferentiated Meloidogyne incognita eggs leads to irregular growth and embryonic lethality[J]. Int J Parasitol, 2010, 40(11):1303-1310.
doi: 10.1016/j.ijpara.2010.03.010 URL |
| [10] |
Fan WJ, Wei ZR, Zhang M, et al. Resistance to Ditylenchus destructor infection in sweet potato by the expression of small interfering RNAs targeting unc-15, a movement-related gene[J]. Phytopathology, 2015, 105(11):1458-1465.
doi: 10.1094/PHYTO-04-15-0087-R URL |
| [11] |
Niu JH, Jian H, Xu JM, et al. RNAi silencing of the Meloidogyne incognita Rpn7 gene reduces nematode parasitic success[J]. Eur J Plant Pathol, 2012, 134(1):131-144.
doi: 10.1007/s10658-012-9971-y URL |
| [12] |
Joshi I, Kumar A, Kohli D, et al. Conferring root-knot nematode resistance via host-delivered RNAi-mediated silencing of four Mi-msp genes in Arabidopsis[J]. Plant Sci, 2020, 298:110592.
doi: 10.1016/j.plantsci.2020.110592 pmid: 32771150 |
| [13] |
Klink VP, Kim KH, Martins V, et al. A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max[J]. Planta, 2009, 230(1):53-71.
doi: 10.1007/s00425-009-0926-2 URL |
| [14] | 梅眉, 黄永红, 茆振川, 等. 南方根结线虫乳酸脱氢酶基因克隆及其沉默效应分析[J]. 中国农业科学, 2013, 46(16):3384-3391. |
| Mei M, Huang YH, Mao ZC, et al. Molecular clone and its RNA interference effect analysis of lactate dehydrogenase gene in Meloidogyne incognita[J]. Sci Agric Sin, 2013, 46(16):3384-3391. | |
| [15] |
Huang YH, Mei M, Mao ZC, et al. Molecular cloning and virus-induced gene silencing of MiASB in the southern root-knot nematode, Meloidogyne incognita[J]. Eur J Plant Pathol, 2014, 138(1):181-193.
doi: 10.1007/s10658-013-0321-5 URL |
| [16] |
Banerjee S, Banerjee A, Gill SS, et al. RNA interference:a novel source of resistance to combat plant parasitic nematodes[J]. Front Plant Sci, 2017, 8:834.
doi: 10.3389/fpls.2017.00834 pmid: 28580003 |
| [17] |
Kim YH, Yang JW. Recent research on enhanced resistance to parasitic nematodes in sweetpotato[J]. Plant Biotechnol Rep, 2019, 13(6):559-566.
doi: 10.1007/s11816-019-00557-w URL |
| [18] |
Pintard L, Willis JH, Willems A, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase[J]. Nature, 2003, 425(6955):311-316.
doi: 10.1038/nature01959 URL |
| [19] |
Luke-Glaser S, Pintard L, Lu CG, et al. The BTB protein MEL-26 promotes cytokinesis in C. elegans by a CUL-3-independent mechanism[J]. Curr Biol, 2005, 15(18):1605-1615.
pmid: 16169482 |
| [20] | Wilson KJ, Qadota H, Mains PE, et al. UNC-89(obscurin)binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1(katanin)in striated muscle of Caenorhabditis elegans[J]. Mol Biol Cell, 2012, 23(14):2623-2634. |
| [21] |
Quintin S, Mains PE, Zinke A, et al. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo[J]. EMBO Rep, 2003, 4(12):1175-1181.
pmid: 14634695 |
| [22] |
Simmer F, Moorman C, van der Linden AM, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions[J]. PLoS Biol, 2003, 1(1):e12. DOI: 10.1371/journal.pbio.0000012.
doi: 10.1371/journal.pbio.0000012 URL |
| [23] |
Fraser AG, Kamath RS, Zipperlen P, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference[J]. Nature, 2000, 408(6810):325-330.
doi: 10.1038/35042517 URL |
| [24] |
Sönnichsen B, Koski LB, Walsh A, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans[J]. Nature, 2005, 434(7032):462-469.
doi: 10.1038/nature03353 URL |
| [25] |
Lu CG, Mains PE. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis[J]. Dev Biol, 2007, 302(2):438-447.
doi: 10.1016/j.ydbio.2006.09.053 URL |
| [26] |
Johnson JL, Lu C, Raharjo E, et al. Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis[J]. Dev Biol, 2009, 330(2):349-357.
doi: 10.1016/j.ydbio.2009.04.004 URL |
| [27] |
Elvin M, Snoek LB, Frejno M, et al. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes[J]. BMC Genomics, 2011, 12:510.
doi: 10.1186/1471-2164-12-510 pmid: 22004469 |
| [28] | Young HK, Seung HO. In vitro culture and factors affecting population changes of Ditylenchus destructor of ginseng[J]. Korean J Plant Pathol, 1995, 11(1):39-46. |
| [29] |
Scotto-Lavino E, Du G, Frohman MA. 3’ end cDNA amplification using classic RACE[J]. Nat Protoc, 2006, 1(6):2742-2745.
pmid: 17406530 |
| [30] |
Scotto-Lavino E, Du G, Frohman MA. 5’ end cDNA amplification using classic RACE[J]. Nat Protoc, 2006, 1(6):2555-2562.
pmid: 17406509 |
| [31] | 丁中, 彭德良, 何旭峰, 等. 不同地理种群甘薯茎线虫对不同类型杀线剂的敏感性[J]. 农药, 2007, 46(12):851-853. |
| Ding Z, Peng DL, He XF, et al. Susceptibility of different populations of Ditylenchus destructor to different type of nematicides in Hebei Province of China[J]. Agrochemicals, 2007, 46(12):851-853. | |
| [32] |
Perry R, Warrior P, Twomey U, et al. Effects of the biological nematicide, DiTera®, on movement and sensory responses of second stage juveniles of Globodera rostochiensis, and stylet activity of G. rostochiensis and fourth stage juveniles of Ditylenchus dipsaci[J]. Nematology, 2002, 4(8):909-915.
doi: 10.1163/156854102321122520 URL |
| [33] |
Hennessy J, Clarke AJ. Movement of Globodera rostochiensis(wollenweber)juveniles stimulated by potato-root exudate[J]. Nematologica, 1984, 30(2):206-212.
doi: 10.1163/187529284X00103 URL |
| [34] | Blumenthal T, Steward K. RNA processing and gene structure[M]//Riddle DL, Blumenthal T, Meyer BJ, et al. C elegans II. Cold Spring Harbor(NY);Cold Spring Harbor Laboratory Press, 1997:117-45. |
| [35] |
Conrad R, Fen Liou R, Blumenthal T. Functional analysis of a C. elegans trans-splice acceptor[J]. Nucl Acids Res, 1993, 21(4):913-919.
doi: 10.1093/nar/21.4.913 URL |
| [36] |
Zhang H, Blumenthal T. Functional analysis of an intron 3’ splice site in Caenorhabditis elegans[J]. RNA, 1996, 2(4):380-388.
pmid: 8634918 |
| [37] |
Ogg SC, Anderson P, Wickens MP. Splicing of a C. elegans myosin pre-mRNA in a human nuclear extract[J]. Nucl Acids Res, 1990, 18(1):143-149.
doi: 10.1093/nar/18.1.143 URL |
| [38] |
Gorlova O, Fedorov A, Logothetis C, et al. Genes with a large intronic burden show greater evolutionary conservation on the protein level[J]. BMC Evol Biol, 2014, 14(1):50.
doi: 10.1186/1471-2148-14-50 pmid: 24629165 |
| [39] |
Dow MR, Mains PE. Genetic and molecular characterization of the Caenorhabditis elegans gene, mel-26, a postmeiotic negative regulator of Mei-1, a meiotic-specific spindle component[J]. Genetics, 1998, 150(1):119-128.
pmid: 9725834 |
| [40] |
Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants[J]. Genome Res, 2006, 16(8):1017-1030.
pmid: 16825662 |
| [41] |
Xu L, Wei Y, Reboul J, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3[J]. Nature, 2003, 425(6955):316-321.
doi: 10.1038/nature01985 URL |
| [42] |
Weber H, Bernhardt A, Dieterle M, et al. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family[J]. Plant Physiol, 2005, 137(1):83-93.
doi: 10.1104/pp.104.052654 URL |
| [43] |
Zhuang M, Calabrese MF, Liu J, et al. Structures of SPOP-substrate complexes:insights into molecular architectures of BTB-Cul3 ubiquitin ligases[J]. Mol Cell, 2009, 36(1):39-50.
doi: 10.1016/j.molcel.2009.09.022 pmid: 19818708 |
| [44] |
Chen J, Ou Y, Yang Y, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing[J]. Nature, 2018, 557(7706):585-589.
doi: 10.1038/s41586-018-0128-9 URL |
| [45] |
Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes:a summary of the current status[J]. Parasitology, 2012, 139(5):630-640.
doi: 10.1017/S0031182011002071 pmid: 22217302 |
| [46] |
Li WS, Gao BX, Lee SM, et al. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination[J]. Dev Cell, 2007, 12(2):235-246.
doi: 10.1016/j.devcel.2006.12.002 URL |
| [1] | NI Chun-hui, LI Hui-xia, LI Wen-hao, LIU Yong-gang, XU Xue-fen, HAN Bian. Comparison of Molecular Characteristics of the Hybrid Progenies from Different Haplotypes of Ditylenchus destructor [J]. Biotechnology Bulletin, 2021, 37(7): 118-126. |
| [2] | ZHAO Hong-hai, LIANG Chen, ZHANG Yu, DUAN Fang-meng, SONG Wen-wen, SHI Qian-qian, HUANG Wen-kun, PENG De-liang. Research Advances of Biology in Ditylenchus destructor Thorne,1945 [J]. Biotechnology Bulletin, 2021, 37(7): 45-55. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||