








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (7): 137-145.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0010
Previous Articles Next Articles
LIU Juan( ), ZHU Chun-xiao, XIAO Xue-qiong, MO Chen-mi, WANG Gao-feng(
), ZHU Chun-xiao, XIAO Xue-qiong, MO Chen-mi, WANG Gao-feng( ), XIAO Yan-nong(
), XIAO Yan-nong( )
)
Received:2021-01-04
Online:2021-07-26
Published:2021-08-13
Contact:
WANG Gao-feng,XIAO Yan-nong
E-mail:juandalin0704@163.com;jksgo@mail.hzau.edu.cn;xiaoyannong@mail.hzau.edu.cn
LIU Juan, ZHU Chun-xiao, XIAO Xue-qiong, MO Chen-mi, WANG Gao-feng, XIAO Yan-nong. Screening of Protein Interacting with Purpureocillium lilacinum Cyclophilin PlCYP6[J]. Biotechnology Bulletin, 2021, 37(7): 137-145.
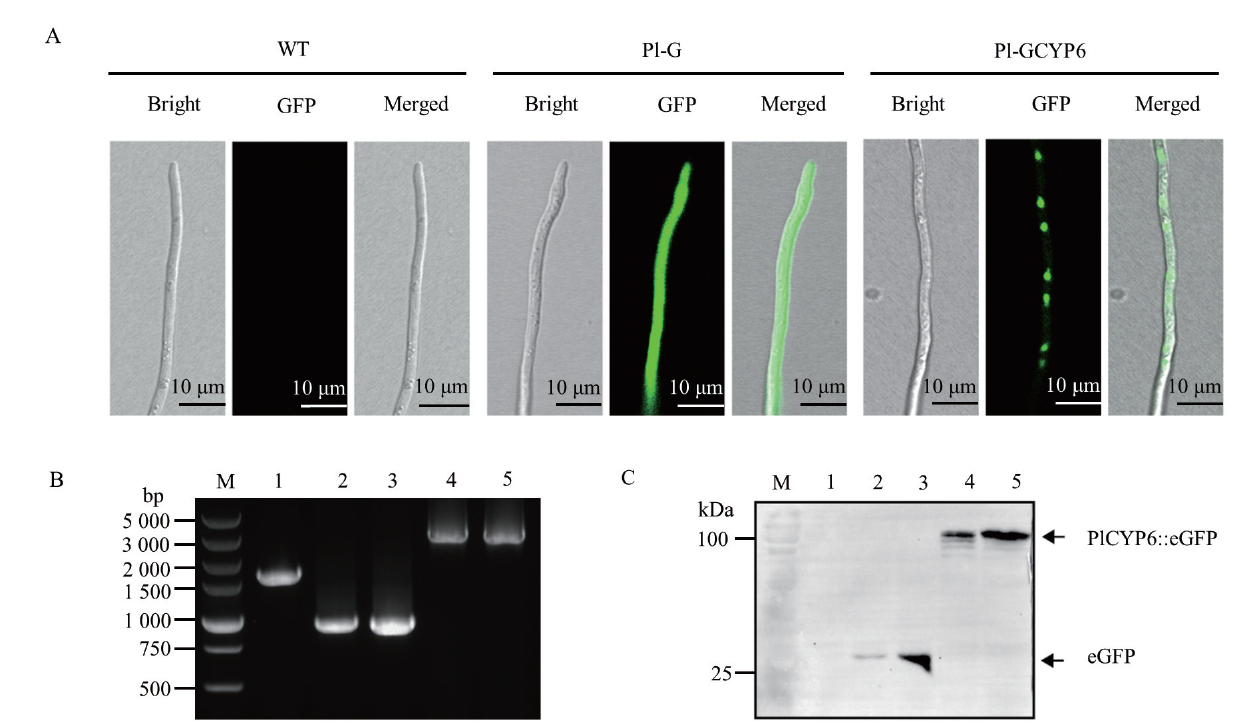
Fig. 1 Heterologous expressions of eGFP and PlCYP6∷eGFP in P. lilacinum A:Observation of the eGFP green fluorescence signal of P. lilacinum(WT),strains Pl-G(heterologous expression of eGFP)and Pl-GCYP6(heterologous expression of PlCYP6∷eGFP)by confocal microscope. B:Amplification of eGFP(about 1 000 bp)and PlCYP6∷eGFP(about 3 000 bp)by PCR,lane M is a DNA marker;lane 1 is WT;lane 2 and lane 3 are strain Pl-G;lane 4 and lane 5 are the strain Pl-GCYP6. C:Detection of eGFP or PlCYP6∷eGFP in the total protein of P. lilacinum by Western blot. Among them,using eGFP-anti as the primary antibody and goat anti-mouse IgG as the secondary antibody;lane M is Precision Plus ProteinTM Standards,lane 1 is the total protein of WT;lane 2 and lane 3 are the total protein of strain Pl-G;lane 4 and lane 5 are the total protein of strain Pl-GCYP6
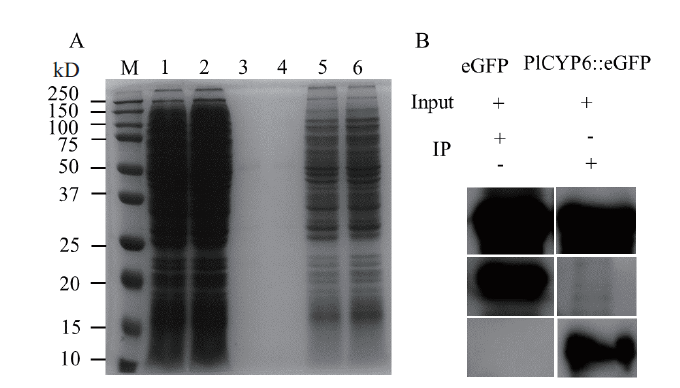
Fig. 2 Immunoprecipitation to catch interacting proteins of PlCYP6 A:Detection of proteins by SDS-PAGE gel electrophoresis;M is Precision Plus ProteinTM Standards;lane 1 and lane 2 are the extracted total protein of Pl-G strain(heterologous expression of eGFP)and Pl-GCYP6 strain(heterologous expression of PlCYP6∷eGFP)after treatment with 1 mol/L NaCl for 0,15,30 and 60 min;lane 3 and lane 4 are the eluted products of total protein after immunoprecipitation;lane 5 and lane 6 are supernatant that washed for the first time when non-specific proteins were removed after immunoprecipitation. B:Western blot was used to hybridize the eluted product after immunoprecipitation;Input was the total protein solution;eGFP-anti as the primary antibody and goat anti-mouse IgG as the secondary antibody;“+” indicates containing and “-” indicates not containing

Fig. 3 Distribution of differential proteins Control group:Pl-G strain(heterologous expression of eGFP). Treatment group:Pl-GCYP6 strain(heterologous expression of PlCYP6∷eGFP). The protein solution after immunoprecipitation in the control group and the treatment group was analyzed by LC-MS/MS to identify the number and types of proteins respectively. The databases are UniProt and the protein databases based on genome annotation. Compared with the control group,the specific proteins in the treatment group were selected as candidate interacting proteins(482)

Fig. 4 GO function annotation and KEGG pathway analysis of candidate interacting proteins Based on the GO database and the KEGG pathway database,the candidate interacting proteins(482)are annotated with protein function. The abscissa is the number of proteins,and the ordinate is the classification item

Fig. 5 Interaction relationship between PlCYP6 and ADH1 protein by yeast two-hybrid A:Principle diagram of truncation analysis based on PlCYP6 protein domain, WD40 repeat and Cyclophilin-like are the predicted protein domains of PlCYP6. B:Yeast two-hybrid self-activation verification. C:Yeast twohybrid results between PlCYP6 and different segmented regions with ADH1. BD:PGBKT7;AD:PGADT7-7;“+” refers to AD or BD empty carrier. If yeast grew on DDO medium(two-deficient medium),the target vectors have been successfully transformed into the yeast. If yeast grew on QDO/X/A medium(four-deficient medium),there will be direct interaction relationship between target proteins

Fig. 6 Analysis of the gene expression levels of PlCYP6 and ADH1 under salt stress The wild-type P.lilacinum was treated with sterile water(Mock)and 1 mol/L NaCl for 60 min,and then the RNA was extracted. Actin was used as an internal control to measure the gene expression levels of PlCYP6 and ADH1. The t test was used to determine significant differences between samples. “***” refers to P<0.001
| [1] |
Zhang L, Yu GX, Xia DW, et al. Protein-protein interactions prediction based on ensemble deep neural networks[J]. Neurocomputing, 2019, 324:10-19.
doi: 10.1016/j.neucom.2018.02.097 |
| [2] | Rao VS, Srinivas K, Sujini GN, et al. Protein-protein interaction detection:methods and analysis[J]. Int J Proteomics, 2014, 2014:147648. |
| [3] |
Huang BX, Kim HY. Effective identification of Akt interacting proteins by two-step chemical crosslinking, co-immunoprecipitation and mass spectrometry[J]. PLoS One, 2013, 8(4):e61430.
doi: 10.1371/journal.pone.0061430 URL |
| [4] |
Li Y, Collins M, An J, et al. Immunoprecipitation and mass spectrometry defines an extensive RBM45 protein-protein interaction network[J]. Brain Res, 2016, 1647:79-93.
doi: 10.1016/j.brainres.2016.02.047 URL |
| [5] |
Phee BK, Shin DH, Cho JH, et al. Identification of phytochrome-interacting protein candidates in Arabidopsis thaliana by co-immunoprecipitation coupled with MALDI-TOF MS[J]. Proteomics, 2006, 6(12):3671-3680.
doi: 10.1002/(ISSN)1615-9861 URL |
| [6] | 李耀东, 关振宏, 严景华. 免疫共沉淀筛选细胞内与A型流感病毒M2蛋白相互作用的蛋白[J]. 微生物学报, 2009, 49(8):1081-1085. |
| Li YD, Guan ZH, Yan JH. Screening cellular proteins interacted with M2 protein of influenza A virus by Coimmunoprecipitation[J]. Acta Microbiol Sin, 2009, 49(8):1081-1085. | |
| [7] | 岳金荣, 丛玉婷, 邢震宇, 等. 利用免疫共沉淀联合质谱技术筛选盐藻MAPK的互作蛋白[J]. 核农学报, 2020, 34(6):1187-1195. |
| Yue JR, Cong YT, Xing ZY, et al. Detection proteins interacting with Dunaliella salina MAPK by co-immunoprecipitation and mass spectrometry[J]. J Nucl Agric Sci, 2020, 34(6):1187-1195. | |
| [8] |
Camilloni C, Sahakyan AB, Holliday MJ, et al. Cyclophilin A catalyzes proline isomerization by an electrostatic handle mechanism[J]. Proc Natl Acad Sci USA, 2014, 111(28):10203-10208.
doi: 10.1073/pnas.1404220111 URL |
| [9] |
Mukherjee D, Patra H, Laskar A, et al. Cyclophilin-mediated reactivation pathway of inactive adenosine kinase aggregates[J]. Arch Biochem Biophys, 2013, 537(1):82-90.
doi: 10.1016/j.abb.2013.06.018 pmid: 23831509 |
| [10] |
Vasudevan D, Fu AG, Luan S, et al. Crystal structure of Arabidopsis Cyclophilin reveals a previously uncharacterized immunophilin fold and a possible autoinhibitory mechanism[J]. Plant Cell, 2012, 24(6):2666-2674.
doi: 10.1105/tpc.111.093781 URL |
| [11] |
Horowitz DS, Lee EJ, Mabon SA, et al. A cyclophilin functions in pre-mRNA splicing[J]. EMBO J, 2002, 21(3):470-480.
pmid: 11823439 |
| [12] |
Li H, Luan S. The cyclophilin AtCYP71 interacts with CAF-1 and LHP1 and functions in multiple chromatin remodeling processes[J]. Mol Plant, 2011, 4(4):748-758.
doi: 10.1093/mp/ssr036 URL |
| [13] |
Li H, He Z, Lu G, et al. A WD40 domain cyclophilin interacts with histone H3 and functions in gene repression and organogenesis in Arabidopsis[J]. Plant Cell, 2007, 19(8):2403-2416.
doi: 10.1105/tpc.107.053579 URL |
| [14] | Hanes SD. Prolyl isomerases in gene transcription[J]. Biochim Biophys Acta, 2015, 1850(10):2017-2034. |
| [15] |
Yurchenko V, O’Connor M, Dai WW, et al. CD147 is a signaling receptor for cyclophilin B[J]. Biochem Biophys Res Commun, 2001, 288(4):786-788.
doi: 10.1006/bbrc.2001.5847 URL |
| [16] |
Brazin KN, Mallis RJ, Fulton DB, et al. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A[J]. Proc Natl Acad Sci USA, 2002, 99(4):1899-1904.
doi: 10.1073/pnas.042529199 URL |
| [17] | Mo CM, Xie C, Wang GF, et al. Genome-wide identification and characterization of the cyclophilin gene family in the nematophagous fungus Purpureocillium lilacinum[J]. Int J Mol Sci, 2019, 20(12):E2978. |
| [18] |
Luangsa-Ard J, Houbraken J, van Doorn T, et al. Purpureocillium, a new genus for the medically important Paecilomyces lilacinus[J]. FEMS Microbiol Lett, 2011, 321(2):141-149.
doi: 10.1111/j.1574-6968.2011.02322.x pmid: 21631575 |
| [19] |
Giné A, Sorribas FJ. Effect of plant resistance and BioAct WG(Purpureocillium lilacinum strain 251)on Meloidogyne incognita in a tomato-cucumber rotation in a greenhouse[J]. Pest Manag Sci, 2017, 73(5):880-887.
doi: 10.1002/ps.2017.73.issue-5 URL |
| [20] |
Wilson MJ, Jackson TA. Progress in the commercialisation of bionematicides[J]. BioControl, 2013, 58(6):715-722.
doi: 10.1007/s10526-013-9511-5 URL |
| [21] | 刘晓艳, 闵勇, 饶犇 等. 杀线虫剂产品研究进展[J]. 中国生物防治学报, 2021, 1-7. |
| Liu XY, Min Y, Rao B, et al. Research Advances on the Nematocides[J]. Chinese Journal of Biological Control, 2021, 1-7. | |
| [22] |
Marian M, Shimizu M. Improving performance of microbial biocontrol agents against plant diseases[J]. J Gen Plant Pathol, 2019, 85(5):329-336.
doi: 10.1007/s10327-019-00866-6 |
| [23] |
Liang LQ, Li JQ, Cheng L, et al. A high efficiency gene disruption strategy using a positive-negative split selection marker and electroporation for Fusarium oxysporum[J]. Microbiol Res, 2014, 169(11):835-843.
doi: 10.1016/j.micres.2014.03.004 URL |
| [24] |
Braun P, Gingras AC. History of protein-protein interactions:from egg-white to complex networks[J]. Proteomics, 2012, 12(10):1478-1498.
doi: 10.1002/pmic.201100563 URL |
| [25] |
De Rybel B, Möller B, Yoshida S, et al. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis[J]. Dev Cell, 2013, 24(4):426-437.
doi: 10.1016/j.devcel.2012.12.013 pmid: 23415953 |
| [26] |
Saiga S, Möller B, Watanabe-Taneda A, et al. Control of embryonic meristem initiation in Arabidopsis by PHD-finger protein complexes[J]. Development, 2012, 139(8):1391-1398.
doi: 10.1242/dev.074492 URL |
| [27] |
Casey JL, Coley AM, Tilley LM, et al. Green fluorescent antibodies:novel in vitro tools[J]. Protein Eng, 2000, 13(6):445-452.
pmid: 10877856 |
| [28] |
Ethier M, Lambert JP, Vasilescu J, et al. Analysis of protein interaction networks using mass spectrometry compatible techniques[J]. Anal Chimica Acta, 2006, 564(1):10-18.
doi: 10.1016/j.aca.2005.12.046 URL |
| [29] |
Rangel DEN. Stress induced cross-protection against environmental challenges on prokaryotic and eukaryotic microbes[J]. World J Microbiol Biotechnol, 2011, 27(6):1281-1296.
doi: 10.1007/s11274-010-0584-3 URL |
| [30] |
Hou Q, Bartels D. Comparative study of the aldehyde dehydrogenase(ALDH)gene superfamily in the glycophyte Arabidopsis thaliana and Eutrema halophytes[J]. Ann Bot, 2015, 115(3):465-479.
doi: 10.1093/aob/mcu152 URL |
| [31] |
Rangel-Porras RA, Meza-Carmen V, Martinez-Cadena G, et al. Molecular analysis of an NAD-dependent alcohol dehydrogenase from the zygomycete Mucor circinelloides[J]. Mol Genet Genomics, 2005, 274(4):354-363.
pmid: 16179992 |
| [32] |
Yi SY, Ku SS, Sim HJ, et al. An alcohol dehydrogenase gene from Synechocystis sp. confers salt tolerance in transgenic tobacco[J]. Front Plant Sci, 2017, 8:1965.
doi: 10.3389/fpls.2017.01965 URL |
| [33] |
Smith TF, Gaitatzes C, Saxena K, et al. The WD repeat:a common architecture for diverse functions[J]. Trends Biochem Sci, 1999, 24(5):181-185.
pmid: 10322433 |
| [34] |
Jain BP, Pandey S. WD40 repeat proteins:signalling scaffold with diverse functions[J]. Protein J, 2018, 37(5):391-406.
doi: 10.1007/s10930-018-9785-7 URL |
| [35] |
Schuetz A, Allali-Hassani A, Martín F, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5[J]. EMBO J, 2006, 25(18):4245-4252.
pmid: 16946699 |
| [1] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [2] | ZHONG Li-ting, CHEN Xiu-zhen, TANG Yun, LI Jun-ren, WANG Xiao-bing, LIU Yan-ting, ZHOU Xuan-xuan, ZHAN Ruo-ting, CHEN Li-kai. Expression of FPPS Recombinant Protein from Pogostemon cablin and Screening of the Interaction Proteins [J]. Biotechnology Bulletin, 2019, 35(12): 10-15. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||