








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 179-186.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0289
Previous Articles Next Articles
WU Kun-kun1,2( ), XU Xing1,2, JI Ce1,2, REN Jian-feng1,2, LI Wei-ming3, ZHANG Qing-hua1,2(
), XU Xing1,2, JI Ce1,2, REN Jian-feng1,2, LI Wei-ming3, ZHANG Qing-hua1,2( )
)
Received:2021-03-10
Online:2022-01-26
Published:2022-02-22
Contact:
ZHANG Qing-hua
E-mail:1017676610@qq.com;qhzhang@shou.edu.cn
WU Kun-kun, XU Xing, JI Ce, REN Jian-feng, LI Wei-ming, ZHANG Qing-hua. Eukaryotic Expression Vector Construction of Danio rerio notch3 Gene and Its Expression Analysis[J]. Biotechnology Bulletin, 2022, 38(1): 179-186.
| 长度 Length/bp | 引物序列 Sequence of primer | 酶切位点选择 Selection of enzyme cutting sites |
|---|---|---|
| 2 487 | TGGGCTAGGCTACTGCTAGTAGG | Pst I和BamH I |
| TCAAGCAAACACCTGCATCTTC |
Table 1 N3ICD primer amplification sequence
| 长度 Length/bp | 引物序列 Sequence of primer | 酶切位点选择 Selection of enzyme cutting sites |
|---|---|---|
| 2 487 | TGGGCTAGGCTACTGCTAGTAGG | Pst I和BamH I |
| TCAAGCAAACACCTGCATCTTC |
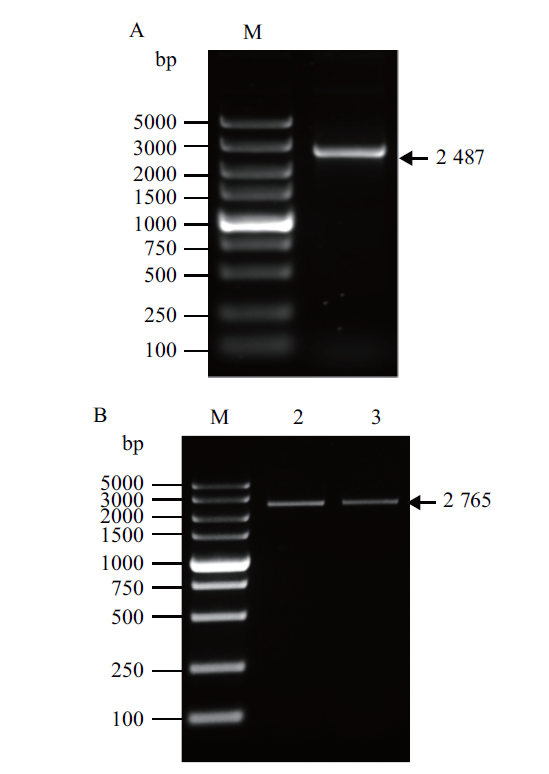
Fig. 2 Cloning of notch3 gene Intracellular domain(A)and construction of recombinant plasmid(B) M:Molecular weight standard. 1:Intracellular segment of notch3 gene. 2, 3:Positive clone
| [1] |
Kopan R, Ilagan MX. The canonical Notch signaling pathway:unfolding the activation mechanism[J]. Cell, 2009, 137(2):216-233.
doi: 10.1016/j.cell.2009.03.045 URL |
| [2] |
Baron M. Combining genetic and biophysical approaches to probe the structure and function relationships of the notch receptor[J]. Mol Membr Biol, 2017, 34(1/2):33-49.
doi: 10.1080/09687688.2018.1503742 URL |
| [3] |
何丽番, 高海. Mecp2在斑马鱼胚胎神经发育中对NOTCH信号通路的调控[J]. 生物技术通报, 2016, 32(4):228-233.
doi: 10.13560/j.cnki.biotech.bull.1985.2016.04.031 |
| He LF, Gao H. The regulation of Mecp2 on notch signaling pathway during early neural development of zebrafish embryos[J]. Biotechnol Bull, 2016, 32(4):228-233. | |
| [4] |
Wharton KA, Johansen KM, Xu T, et al. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats[J]. Cell, 1985, 43(3):567-581.
doi: 10.1016/0092-8674(85)90229-6 URL |
| [5] |
Bellavia D, Checquolo S, Campese AF, et al. Notch3:from subtle structural differences to functional diversity[J]. Oncogene, 2008, 27(38):5092-5098.
doi: 10.1038/onc.2008.230 pmid: 18758477 |
| [6] | Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance[J]. J Cell Sci, 2013, 126(pt 10):2135-2140. |
| [7] | Bray SJ. Notch signalling:a simple pathway becomes complex[J]. Nat Rev Mol Cell Biol, 2006, 7(9):678-689. |
| [8] |
Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system[J]. Immunity, 2010, 32(1):14-27.
doi: 10.1016/j.immuni.2010.01.004 URL |
| [9] |
Xu X, Choi SH, Hu TC, et al. Insights into autoregulation of Notch3 from structural and functional studies of its negative regulatory region[J]. Structure, 2015, 23(7):1227-1235.
doi: 10.1016/j.str.2015.05.001 URL |
| [10] |
Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms[J]. Cell, 1991, 66(4):649-661.
pmid: 1831692 |
| [11] |
Bellavia D, Campese AF, Checquolo S, et al. Combined expression of pTα and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis[J]. PNAS, 2002, 99(6):3788-3793.
pmid: 11891328 |
| [12] |
Franciosa G, Diluvio G, del Gaudio F, et al. Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression[J]. Oncogene, 2016, 35(36):4741-4751.
doi: 10.1038/onc.2016.5 pmid: 26876201 |
| [13] |
Bernasconi-Elias P, Hu T, Jenkins D, et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies[J]. Oncogene, 2016, 35(47):6077-6086.
doi: 10.1038/onc.2016.133 pmid: 27157619 |
| [14] |
Choi SH, Severson E, Pear WS, et al. The common oncogenomic program of NOTCH1 and NOTCH3 signaling in T-cell acute lymphoblastic leukemia[J]. PLoS One, 2017, 12(10):e0185762.
doi: 10.1371/journal.pone.0185762 URL |
| [15] |
Qiao J, Liu J, Jia K, et al. Diosmetin triggers cell apoptosis by activation of the p53/Bcl-2 pathway and inactivation of the Notch3/NF-κB pathway in HepG2 cells[J]. Oncol Lett, 2016, 12(6):5122-5128.
doi: 10.3892/ol.2016.5347 pmid: 28101238 |
| [16] |
Strähle U, Scholz S, Geisler R, et al. Zebrafish embryos as an alternative to animal experiments——a commentary on the definition of the onset of protected life stages in animal welfare regulations[J]. Reprod Toxicol, 2012, 33(2):128-132.
doi: 10.1016/j.reprotox.2011.06.121 pmid: 21726626 |
| [17] | 林金杏, 杨迟, 冯丽萍, 等. 斑马鱼嗜水气单胞菌的鉴定和人工感染组织病理学研究[J]. 生物技术通报, 2016, 32(9):239-245. |
| Lin JX, Yang C, Feng LP, et al. Identification of Aeromonas hydrophila and histopathological observation of artificial infected zebrafish[J]. Biotechnol Bull, 2016, 32(9):239-245. | |
| [18] |
Xie Y, Meijer AH, Schaaf MJM. Modeling inflammation in zebrafish for the development of anti-inflammatory drugs[J]. Front Cell Dev Biol, 2020, 8:620984.
doi: 10.3389/fcell.2020.620984 URL |
| [19] |
Lawrence C, Sanders GE, Varga ZM, et al. Regulatory compliance and the zebrafish[J]. Zebrafish, 2009, 6(4):453-456.
doi: 10.1089/zeb.2009.0595 URL |
| [20] | 张曼, 李均. Notch和TGF信号通路对话在肾间质纤维化的研究[J]. 临床肾脏病杂志, 2019, 19(9):705-709. |
| Zhang M, Li J. A study on the relationship between Notch and TGF signal pathways in renal interstitial fibrosis[J]. J Clin Nephrol, 2019, 19(9):705-709. | |
| [21] | 李明星, 顾胜龙, 应苗法, 等. Notch信号通路在肺动脉高压肺血管重构中的研究进展[J]. 中国药理学通报, 2020, 36(1):5-8. |
| Li MX, Gu SL, Ying MF, et al. Advances in research on Notch signaling pathway in pulmonary vascular remodeling of pulmonary arterial hypertension[J]. Chin Pharmacol Bull, 2020, 36(1):5-8. | |
| [22] | 刘晓聪, 温俊杰, 郭家定, 等. Notch信号通路在肿瘤微环境中的调控作用[J]. 中国肿瘤, 2020, 29(1):48-54. |
| Liu XC, Wen JJ, Guo JD, et al. Regulation of notch signaling pathways in tumor microenvironment[J]. China Cancer, 2020, 29(1):48-54. | |
| [23] |
Natsuizaka M, Whelan KA, Kagawa S, et al. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma[J]. Nat Commun, 2017, 8(1):1758.
doi: 10.1038/s41467-017-01500-9 pmid: 29170450 |
| [24] |
Durrant CS, Ruscher K, Sheppard O, et al. Beta secretase 1-dependent amyloid precursor protein processing promotes excessive vascular sprouting through NOTCH3 signalling[J]. Cell Death Dis, 2020, 11(2):98.
doi: 10.1038/s41419-020-2288-4 pmid: 32029735 |
| [25] |
Hosseini-Alghaderi S, Baron M. Notch3 in development, health and disease[J]. Biomolecules, 2020, 10(3):485.
doi: 10.3390/biom10030485 URL |
| [26] |
Xu HX, Zhu J, Smith S, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization[J]. Nat Immunol, 2012, 13(7):642-650.
doi: 10.1038/ni.2304 URL |
| [27] |
Tsao PN, Wei SC, Huang MT, et al. Lipopolysaccharide-induced Notch signaling activation through JNK-dependent pathway regulates inflammatory response[J]. J Biomed Sci, 2011, 18:56.
doi: 10.1186/1423-0127-18-56 URL |
| [28] |
Varga J, Nicolas A, Petrocelli V, et al. AKT-dependent NOTCH3 activation drives tumor progression in a model of mesenchymal colorectal cancer[J]. J Exp Med, 2020, 217(10):e20191515.
doi: 10.1084/jem.20191515 URL |
| [29] |
López-López S, Monsalve EM, Romero de Ávila MJ, et al. NOTCH3 signaling is essential for NF-κB activation in TLR-activated macrophages[J]. Sci Rep, 2020, 10(1):1-16.
doi: 10.1038/s41598-019-56847-4 URL |
| [1] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [2] | BAO Lin-zhu, SHI Can, LU Ling-er, XU Xing, ZHOU Ze-bin, REN Jian-feng, LI Wei-ming, ZHANG Qing-hua. Regulation of Gene mapk1 in Danio rerio on Gene tp53 [J]. Biotechnology Bulletin, 2021, 37(12): 160-168. |
| [3] | HU Xiao, WANG Bao-bao, DOU Shao-hua, JIANG Nan, FU Chang-zhen, JIN Hang, GAO Feng-shan. Construction of a Eukaryotic Expression Vector of SLA-2 Gene from Yantai Black Pigs and Its Expression [J]. Biotechnology Bulletin, 2021, 37(10): 143-151. |
| [4] | MENG Li, DU Cai-ping. Eukaryotic Expression,Purification and Activity Identification of Rat His-Akt1 Recombinant Protein [J]. Biotechnology Bulletin, 2020, 36(12): 98-103. |
| [5] | WANG Ya-nan, WEN Hai-ruo, WANG Xue. Establishment and Preliminary Exploration of in vitro Pig-a Gene Mutation Assay Based on L5178Y Cells [J]. Biotechnology Bulletin, 2020, 36(1): 220-228. |
| [6] | WANG Mei-lin, NIU Jing-ling, JIN Ruo-qi, XU Ying-ying, CAI Jing-jing, CHEN Qun-li. Eukaryotic Expression Vector Construction of Human RhoB Gene and Its Expression Analysis [J]. Biotechnology Bulletin, 2019, 35(10): 174-179. |
| [7] | ZHAI Xiao-xin, GAO Hua, JIANG Ping, XU Chong-bo, ZHANG Zong-hui, LI Wen-zhe, GAO Feng-shan. Construction of Eukaryotic Expression Vector of SLA-2-HB01 from Hebao Pigs and Its Expression in PK15 Cells [J]. Biotechnology Bulletin, 2018, 34(12): 132-139. |
| [8] | ZHANG Jing-jing, JIN Xiao-bao, LI Xiao-bo, WANG Jie, MA Yan. Overexpression of Glypican-3 and Its Effects on the Biological Behaviors in Highly Metastatic Hepatocarcinoma Cells HCCLM3 [J]. Biotechnology Bulletin, 2017, 33(12): 176-184. |
| [9] | WANG Hai-yan,, ZHU Zhi-xuan, JIN Jing ,DING Yi. The Optimization of Chromosome Preparation and Immunofluorescence Staining for Root Tip of Nelumbo nucifera [J]. Biotechnology Bulletin, 2016, 32(4): 74-79. |
| [10] | Ruan Zheng,Li Jie,Liu Kaiwu,Wang Lianfang,Hua Juan,Hu Xiaoming,Liu Jie,Huang Haijun. Construction of Eukaryotic Expression Vector for the CD151 and CD163 Receptor Proteins of Porcine Reproductive and Respiratory Syndrome Virus [J]. Biotechnology Bulletin, 2014, 0(12): 190-194. |
| [11] | Liu Lili, Wang Jian, Wu Wei, Wang Haisheng, Yan Yanchun. Application of Zebrafish in Research of Transgenic Organisms [J]. Biotechnology Bulletin, 2013, 0(2): 15-21. |
| [12] | Liu Lili Wang Jian Wang Haisheng Yu Kaimin Li Guochao Yan Yanchun . Foundation of Platform for Zebrafish Transgenic Technology [J]. Biotechnology Bulletin, 2013, 0(10): 120-126. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||