








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (4): 184-192.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1198
Previous Articles Next Articles
XIAO Yan-hua1,2( ), ZOU Zhi2(
), ZOU Zhi2( ), ZHAO Yong-guo3, GUO An-ping2, ZHANG Li1(
), ZHAO Yong-guo3, GUO An-ping2, ZHANG Li1( )
)
Received:2021-09-16
Online:2022-04-26
Published:2022-05-06
Contact:
ZOU Zhi, ZHANG Li
E-mail:1740329672@qq.com;zouzhi2008@126.com;zhangli0624@mail.scuec.edu.cn
XIAO Yan-hua, ZOU Zhi, ZHAO Yong-guo, GUO An-ping, ZHANG Li. Molecular Cloning and Characterization of an Acetolactate Synthase Gene(CeALS)from Cyperus esculentus L.[J]. Biotechnology Bulletin, 2022, 38(4): 184-192.

Fig.1 Full-length cDNA sequence of CeALS and its dedu-ced coding amino acids The gene coding region is identified by uppercase base letters,the 5' UTR and 3' UTR are identified by lowercase bases,the cloning primer pair CeALSF/R are marked with single underlines,and the primer pair CeALSHF/R for homologous recombinant was marked by double underlines
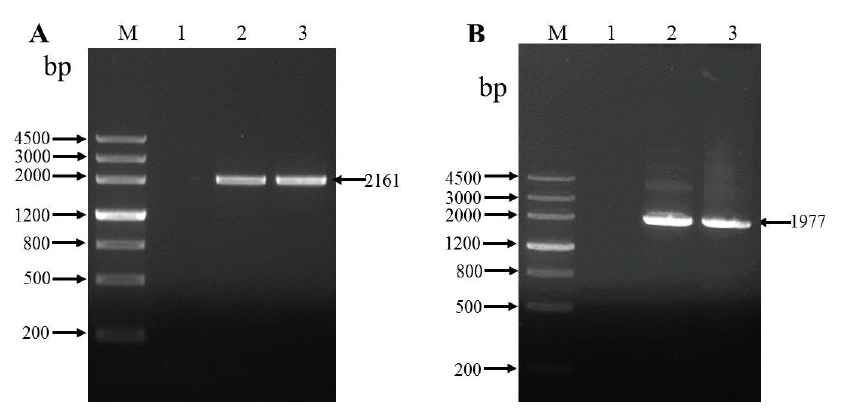
Fig. 2 PCR amplification of CeALS in C. esculentus A:Results of the first round of PCR amplification. B:Results of the second round of PCR amplification. M:DNA marker III. 1:Blank control. 2 and 3:CeALS
| 基因 Gene | 内含子数 Number of intron | 蛋白 Protein/AA | 信号肽 Transit peptide/AA | 分子量 MW/kD | 等电点 pI | 总平均疏水指数 GRAVY | 脂肪族指数 AI | 不稳定系数 II |
|---|---|---|---|---|---|---|---|---|
| AtALS | 0 | 670 | 55 | 72.59 | 6.20 | -0.130 | 87.78 | 42.93 |
| PeALS | 0 | 657 | 58 | 71.51 | 6.78 | -0.126 | 95.16 | 40.20 |
| PdALS | 0 | 658 | 68 | 70.91 | 6.17 | -0.054 | 95.06 | 43.27 |
| EgALS | 0 | 658 | 68 | 70.89 | 6.27 | -0.063 | 94.92 | 41.18 |
| SpALS | 0 | 659 | 52 | 71.81 | 5.97 | -0.138 | 92.67 | 43.44 |
| AcALS | 0 | 655 | 59 | 70.44 | 6.37 | -0.061 | 93.89 | 45.85 |
| MaALS1 | 0 | 655 | 58 | 71.12 | 6.64 | -0.091 | 93.11 | 38.24 |
| MaALS2 | 0 | 660 | 54 | 71.84 | 6.43 | -0.103 | 93.03 | 37.67 |
| OsALS1 | 0 | 644 | 43 | 69.39 | 6.48 | -0.027 | 92.13 | 41.61 |
| OsALS2 | 0 | 663 | 79 | 71.18 | 6.30 | -0.111 | 87.59 | 36.30 |
| HvALS | 0 | 646 | 46 | 69.35 | 6.58 | -0.004 | 92.32 | 40.49 |
| BdALS1 | 0 | 646 | 46 | 69.45 | 6.55 | -0.015 | 92.46 | 43.17 |
| BdALS2 | 0 | 646 | 46 | 70.12 | 6.41 | -0.066 | 92.15 | 37.31 |
| SvALS | 0 | 643 | 41 | 69.39 | 6.45 | -0.053 | 88.96 | 42.81 |
| SiALS | 0 | 643 | 41 | 69.38 | 6.45 | -0.054 | 88.96 | 42.84 |
| SbALS | 0 | 641 | 40 | 69.05 | 6.42 | 0.000 | 92.14 | 40.01 |
| ZmALS1 | 0 | 638 | 39 | 68.93 | 6.69 | -0.060 | 91.16 | 40.26 |
| ZmALS2 | 0 | 638 | 39 | 68.99 | 6.77 | -0.036 | 91.33 | 40.15 |
| ClALS1 | 1 | 644 | 79 | 70.11 | 6.45 | -0.145 | 89.35 | 38.09 |
| ClALS2 | 1 | 644 | 79 | 70.34 | 6.80 | -0.163 | 88.59 | 37.94 |
| CbALS | 1 | 647 | 55 | 70.46 | 5.85 | -0.166 | 88.92 | 46.54 |
| CeALS | 1 | 645 | 54 | 69.94 | 6.10 | -0.156 | 89.36 | 44.93 |
| SwALS1 | 1 | 645 | 80 | 70.24 | 6.24 | -0.144 | 90.59 | 41.67 |
| SwALS2 | 1 | 645 | 77 | 70.26 | 6.45 | -0.163 | 89.98 | 41.97 |
| SjALS1 | 1 | 645 | 80 | 70.22 | 6.24 | -0.140 | 90.74 | 41.67 |
| SjALS2 | 1 | 645 | 80 | 70.26 | 6.45 | -0.163 | 89.98 | 41.97 |
| SmALS1 | 1 | 648 | 80 | 70.58 | 6.22 | -0.143 | 90.62 | 41.68 |
| SmALS2 | 1 | 645 | 77 | 70.21 | 6.45 | -0.157 | 90.28 | 41.37 |
Table 1 Gene structure and physicochemical properties of ALS genes in Arabidopsis and 20 monocots
| 基因 Gene | 内含子数 Number of intron | 蛋白 Protein/AA | 信号肽 Transit peptide/AA | 分子量 MW/kD | 等电点 pI | 总平均疏水指数 GRAVY | 脂肪族指数 AI | 不稳定系数 II |
|---|---|---|---|---|---|---|---|---|
| AtALS | 0 | 670 | 55 | 72.59 | 6.20 | -0.130 | 87.78 | 42.93 |
| PeALS | 0 | 657 | 58 | 71.51 | 6.78 | -0.126 | 95.16 | 40.20 |
| PdALS | 0 | 658 | 68 | 70.91 | 6.17 | -0.054 | 95.06 | 43.27 |
| EgALS | 0 | 658 | 68 | 70.89 | 6.27 | -0.063 | 94.92 | 41.18 |
| SpALS | 0 | 659 | 52 | 71.81 | 5.97 | -0.138 | 92.67 | 43.44 |
| AcALS | 0 | 655 | 59 | 70.44 | 6.37 | -0.061 | 93.89 | 45.85 |
| MaALS1 | 0 | 655 | 58 | 71.12 | 6.64 | -0.091 | 93.11 | 38.24 |
| MaALS2 | 0 | 660 | 54 | 71.84 | 6.43 | -0.103 | 93.03 | 37.67 |
| OsALS1 | 0 | 644 | 43 | 69.39 | 6.48 | -0.027 | 92.13 | 41.61 |
| OsALS2 | 0 | 663 | 79 | 71.18 | 6.30 | -0.111 | 87.59 | 36.30 |
| HvALS | 0 | 646 | 46 | 69.35 | 6.58 | -0.004 | 92.32 | 40.49 |
| BdALS1 | 0 | 646 | 46 | 69.45 | 6.55 | -0.015 | 92.46 | 43.17 |
| BdALS2 | 0 | 646 | 46 | 70.12 | 6.41 | -0.066 | 92.15 | 37.31 |
| SvALS | 0 | 643 | 41 | 69.39 | 6.45 | -0.053 | 88.96 | 42.81 |
| SiALS | 0 | 643 | 41 | 69.38 | 6.45 | -0.054 | 88.96 | 42.84 |
| SbALS | 0 | 641 | 40 | 69.05 | 6.42 | 0.000 | 92.14 | 40.01 |
| ZmALS1 | 0 | 638 | 39 | 68.93 | 6.69 | -0.060 | 91.16 | 40.26 |
| ZmALS2 | 0 | 638 | 39 | 68.99 | 6.77 | -0.036 | 91.33 | 40.15 |
| ClALS1 | 1 | 644 | 79 | 70.11 | 6.45 | -0.145 | 89.35 | 38.09 |
| ClALS2 | 1 | 644 | 79 | 70.34 | 6.80 | -0.163 | 88.59 | 37.94 |
| CbALS | 1 | 647 | 55 | 70.46 | 5.85 | -0.166 | 88.92 | 46.54 |
| CeALS | 1 | 645 | 54 | 69.94 | 6.10 | -0.156 | 89.36 | 44.93 |
| SwALS1 | 1 | 645 | 80 | 70.24 | 6.24 | -0.144 | 90.59 | 41.67 |
| SwALS2 | 1 | 645 | 77 | 70.26 | 6.45 | -0.163 | 89.98 | 41.97 |
| SjALS1 | 1 | 645 | 80 | 70.22 | 6.24 | -0.140 | 90.74 | 41.67 |
| SjALS2 | 1 | 645 | 80 | 70.26 | 6.45 | -0.163 | 89.98 | 41.97 |
| SmALS1 | 1 | 648 | 80 | 70.58 | 6.22 | -0.143 | 90.62 | 41.68 |
| SmALS2 | 1 | 645 | 77 | 70.21 | 6.45 | -0.157 | 90.28 | 41.37 |

Fig. 3 Multiple sequence alignment and sequence features of CeALS protein The acetolactate synthase domain is marked by solid arrows at both ends. Chloroplast transit peptide and domains of TPP_enzyme_N,TPP_enzyme_M,and TPP_enzyme_C are identified by wire frames,whereas conserved residues that can cause resistance changes are identified by hollow arrows
| 位点Site | CeALS | SNP | 位点Site | CeALS | SNP | 位点Site | CeALS | SNP | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 120 | A | C | 435 | C | G | 1431 | T | G | |||
| 147 | C | T | 519 | C | T | 1512 | T | A | |||
| 237 | G | A | 631 | A | C | 1596 | T | C | |||
| 255 | G | A | 681 | T | C | 1612 | T | G | |||
| 405 | A | G | 685 | C | A | 1702 | A | G | |||
| 408 | T | C | 840 | C | T | 1749 | T | C | |||
| 411 | C | G | 945 | C | T | 1752 | T | C | |||
| 414 | C | T | 1128 | C | T | 1767 | T | C | |||
| 417 | C | T | 1146 | G | A | 1771 | G | T | |||
| 420 | C | A | 1404 | G | A | 1851 | G | T |
Table 2 Distribution of SNPs
| 位点Site | CeALS | SNP | 位点Site | CeALS | SNP | 位点Site | CeALS | SNP | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 120 | A | C | 435 | C | G | 1431 | T | G | |||
| 147 | C | T | 519 | C | T | 1512 | T | A | |||
| 237 | G | A | 631 | A | C | 1596 | T | C | |||
| 255 | G | A | 681 | T | C | 1612 | T | G | |||
| 405 | A | G | 685 | C | A | 1702 | A | G | |||
| 408 | T | C | 840 | C | T | 1749 | T | C | |||
| 411 | C | G | 945 | C | T | 1752 | T | C | |||
| 414 | C | T | 1128 | C | T | 1767 | T | C | |||
| 417 | C | T | 1146 | G | A | 1771 | G | T | |||
| 420 | C | A | 1404 | G | A | 1851 | G | T |
| [1] | 李杨, 马智宏, 李冰茹, 等. 我国主要作物中除草剂登记情况及存在问题[J]. 食品安全质量检测学报, 2018, 9(17):4483-4488. |
| Li Y, Ma ZH, Li BR, et al. Registration status and existing problems of herbicides in the major crops in China[J]. J Food Saf Qual, 2018, 9(17):4483-4488. | |
| [2] | 张玉池, 王晓蕾, 徐文蓉, 等. 国内外抗除草剂基因专利的分析[J]. 杂草学报, 2017, 35(2):1-22. |
| Zhang YC, Wang XL, Xu WR, et al. Analysis on the patents of herbicide resistance gene at home and abroad[J]. J Weed Sci, 2017, 35(2):1-22. | |
| [3] |
Liu YD, Li YY, Wang XY. Acetohydroxyacid synthases:evolution, structure, and function[J]. Appl Microbiol Biotechnol, 2016, 100(20):8633-8649.
doi: 10.1007/s00253-016-7809-9 URL |
| [4] |
Yu Q, Powles SB. Resistance to AHAS inhibitor herbicides:current understanding[J]. Pest Manag Sci, 2014, 70(9):1340-1350.
doi: 10.1002/ps.3710 URL |
| [5] | Heap I. The International Herbicide-Resistant Weed Database[EB/OL]. 2021 www.weedscience.org. |
| [6] | 张学昆. 我国油莎豆产业研发进展报告[J]. 中国农村科技, 2019(4):67-69. |
| Zhang XK. Research progress of the tigernut industry in China[J]. China Rural Sci Technol, 2019(4):67-69. | |
| [7] |
Turesson H, Marttila S, Gustavsson KE, et al. Characterization of oil and starch accumulation in tubers of Cyperus esculentus var. sativus(Cyperaceae):a novel model system to study oil reserves in nonseed tissues[J]. Am J Bot, 2010, 97(11):1884-1893.
doi: 10.3732/ajb.1000200 pmid: 21616827 |
| [8] | 邹智, 赵永国, 张丽, 等. 基于单分子实时测序的油莎豆全长转录组分析[J]. 中国油料作物学报, 2021, 43(2):229-235. |
| Zou Z, Zhao YG, Zhang L, et al. Single-molecule real-time(SMRT)-based full-length transcriptome analysis of tigernut(Cyperus esculentus L.)[J]. Chin J Oil Crop Sci, 2021, 43(2):229-235. | |
| [9] |
De Castro O, Gargiulo R, Del Guacchio E, et al. A molecular survey concerning the origin of Cyperus esculentus(Cyperaceae, Poales):two sides of the same coin(weed vs. crop)[J]. Ann Bot, 2015, 115(5):733-745.
doi: 10.1093/aob/mcv001 URL |
| [10] |
Tehranchian P, Norsworthy JK, Nandula V, et al. First report of resistance to acetolactate-synthase-inhibiting herbicides in yellow nutsedge(Cyperus esculentus):confirmation and characterization[J]. Pest Manag Sci, 2015, 71(9):1274-1280.
doi: 10.1002/ps.3922 pmid: 25307777 |
| [11] | Okuno J, Iwakami S, Uchino A, et al. Responses to halosulfuron-methyl and flazasulfuron and mutation of acetolactate synthase gene of Cyperus brebifolius survived in turf grass on golf course[J]. J Jpn Soc Turfgrass Sci, 2015, 43:159-162. |
| [12] |
Ntoanidou S, Kaloumenos N, Diamantidis G, et al. Molecular basis of Cyperus difformis cross-resistance to ALS-inhibiting herbicides[J]. Pestic Biochem Physiol, 2016, 127:38-45.
doi: 10.1016/j.pestbp.2015.09.004 pmid: 26821656 |
| [13] |
Goodstein DM, Shu S, Howson R, et al. Phytozome:a comparative platform for green plant genomics[J]. Nucleic Acids Res, 2012, 40(database issue):D1178-D1186.
doi: 10.1093/nar/gkr944 URL |
| [14] |
Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank[J]. Nucleic Acids Res, 2010, 38(database issue):D46-D51.
doi: 10.1093/nar/gkp1024 URL |
| [15] | 邹智, 杨礼富. 巴西橡胶树铁硫簇支架蛋白cDNA的克隆与分析[J]. 热带作物学报, 2010, 31(10):1752-1756. |
| Zou Z, Yang LF. Cloning and analysis of a cDNA encoding a iron-sulfur cluster scaffold protein ISU1 from Hevea brasiliensis Müll. Arg[J]. Chin J Trop Crops, 2010, 31(10):1752-1756. | |
| [16] | 赵永国, 危文亮. 利用SRAP标记分析油莎豆遗传多样性[J]. 中国油料作物学报, 2011, 33(4):351-355. |
| Zhao YG, Wei WL. Genetic diversity analysis of tigernut(Cyperus esculentus)using SRAP markers[J]. Chin J Oil Crop Sci, 2011, 33(4):351-355. | |
| [17] |
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2010, 26(5):589-595.
doi: 10.1093/bioinformatics/btp698 URL |
| [18] |
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit:a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Res, 2010, 20(9):1297-1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [19] |
Zou Z, Xie GS, Yang LF. Papain-like cysteine protease encoding genes in rubber(Hevea brasiliensis):comparative genomics, phylogenetic, and transcriptional profiling analysis[J]. Planta, 2017, 246(5):999-1018.
doi: 10.1007/s00425-017-2739-z URL |
| [20] |
Zou Z, Gong J, An F, et al. Genome-wide identification of rubber tree(Hevea brasiliensis Muell. Arg. )aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue[J]. BMC Genom, 2015, 16(1):1-18.
doi: 10.1186/1471-2164-16-1 URL |
| [21] |
Uchino A, Ogata S, Kohara H, et al. Molecular basis of diverse responses to acetolactate synthase-inhibiting herbicides in sulfonylurea-resistant biotypes of Schoenoplectus juncoides[J]. Weed Biol Manage, 2007, 7(2):89-96.
doi: 10.1111/j.1445-6664.2007.00240.x URL |
| [22] |
Scarabel L, Locascio A, Furini A, et al. Characterisation of ALS genes in the polyploid species Schoenoplectus mucronatus and implications for resistance management[J]. Pest Manag Sci, 2010, 66(3):337-344.
doi: 10.1002/ps.1883 pmid: 19921713 |
| [23] |
Sada Y, Ikeda H, Yamato S, et al. Characterization of sulfonylurea-resistant Schoenoplectus juncoides having a target-site Asp(376)Glu mutation in the acetolactate synthase[J]. Pestic Biochem Physiol, 2013, 107(1):106-111.
doi: 10.1016/j.pestbp.2013.05.013 URL |
| [24] |
Riar DS, Tehranchian P, Norsworthy JK, et al. Acetolactate synthase-inhibiting, herbicide-resistant rice flatsedge(Cyperus iria):cross-resistance and molecular mechanism of resistance[J]. Weed Sci, 2015, 63(4):748-757.
doi: 10.1614/WS-D-15-00014.1 URL |
| [25] |
McCullough PE, Yu JL, McElroy JS, et al. ALS-resistant annual sedge(Cyperus compressus)confirmed in turf grass[J]. Weed Sci, 2016, 64(1):33-41.
doi: 10.1614/WS-D-15-00094.1 URL |
| [26] |
Larridon I, Reynders M, Huygh W, et al. Affinities in C3 Cyperus li-neages(Cyperaceae)revealed using molecular phylogenetic data and carbon isotope analysis[J]. Bot J Linn Soc, 2011, 167(1):19-46.
doi: 10.1111/j.1095-8339.2011.01160.x URL |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||