








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (8): 60-68.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1353
Previous Articles Next Articles
GAO Wei-xin( ), HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng(
), HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng( )
)
Received:2021-10-29
Online:2022-08-26
Published:2022-09-14
Contact:
YANG Hao-meng
E-mail:gaoweixin19@163.com;yhmbjbj@126.com
GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing[J]. Biotechnology Bulletin, 2022, 38(8): 60-68.
| Primer name | Primer sequence(5'-3') |
|---|---|
| Pet-Insert-F | taagaaggagatataccatggATAAGAAATACTCAATAGGCTTAGA |
| Pet-Insert-R | gtggtggtggtggtgctcgagGACCTTGCGCTTCTTCTTGGgagggtcacctcctagctgactcaaatc |
| glucoamylase-F | atgtcgttccgatctctactcgc |
| glucoamylase-R | ctaccgccaggtgtcagtcac |
| 223up-F | atgtcgttccgatctctactcgccctg |
| 223up-R | acttcttcccagagatctgcgatcg |
| 223down-F | caatggctcgtctttctttacgattgc |
| 223down-R | aggtgacagtcacactgctgtagg |
| hph-F | gcagatctctgggaagaagtgacagaagatgatattgaaggagcac |
| hph-R | aaagaaagacgagccattggtatctggaagaggtaaacccgaaacg |
| hph-jc-F | ctcgccgatagtggaaaccgac |
| hph-jc-R | cgcaccaagttatcgtgcacc |
| GA223homo-up-jc-F | cccacaccatcggagattcgtcgcc |
Table 1 Primer names and sequence information
| Primer name | Primer sequence(5'-3') |
|---|---|
| Pet-Insert-F | taagaaggagatataccatggATAAGAAATACTCAATAGGCTTAGA |
| Pet-Insert-R | gtggtggtggtggtgctcgagGACCTTGCGCTTCTTCTTGGgagggtcacctcctagctgactcaaatc |
| glucoamylase-F | atgtcgttccgatctctactcgc |
| glucoamylase-R | ctaccgccaggtgtcagtcac |
| 223up-F | atgtcgttccgatctctactcgccctg |
| 223up-R | acttcttcccagagatctgcgatcg |
| 223down-F | caatggctcgtctttctttacgattgc |
| 223down-R | aggtgacagtcacactgctgtagg |
| hph-F | gcagatctctgggaagaagtgacagaagatgatattgaaggagcac |
| hph-R | aaagaaagacgagccattggtatctggaagaggtaaacccgaaacg |
| hph-jc-F | ctcgccgatagtggaaaccgac |
| hph-jc-R | cgcaccaagttatcgtgcacc |
| GA223homo-up-jc-F | cccacaccatcggagattcgtcgcc |
| Name | Sequence(5'-3') |
|---|---|
| gRNA | TAATACGACTCACTATAGGgatctctgggaagaagtcaa GTTTTAGAGCTAGAAATAGC AAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT |
| cgRNA | TAATACGACTCACTATAGGgatgggattcgtgagtacaaGTTTTAGAGCTAGAAATAGC AAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT |
Table 2 Sequence information of in vitro transcription templates of RNA
| Name | Sequence(5'-3') |
|---|---|
| gRNA | TAATACGACTCACTATAGGgatctctgggaagaagtcaa GTTTTAGAGCTAGAAATAGC AAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT |
| cgRNA | TAATACGACTCACTATAGGgatgggattcgtgagtacaaGTTTTAGAGCTAGAAATAGC AAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT |
| 添加量Additive amount | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Cas9 | 200 ng | 400 ng | 800 ng | 1.6 μg | 3.2 μg | 6.4 μg |
| gRNA | 200 ng | 400 ng | 800 ng | 1.6 μg | 3.2 μg | 6.4 μg |
| DNA | 200 ng | 200 ng | 200 ng | 200 ng | 200 ng | 200 ng |
Table 3 Reaction system of test group for RNP complex activity verification
| 添加量Additive amount | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Cas9 | 200 ng | 400 ng | 800 ng | 1.6 μg | 3.2 μg | 6.4 μg |
| gRNA | 200 ng | 400 ng | 800 ng | 1.6 μg | 3.2 μg | 6.4 μg |
| DNA | 200 ng | 200 ng | 200 ng | 200 ng | 200 ng | 200 ng |
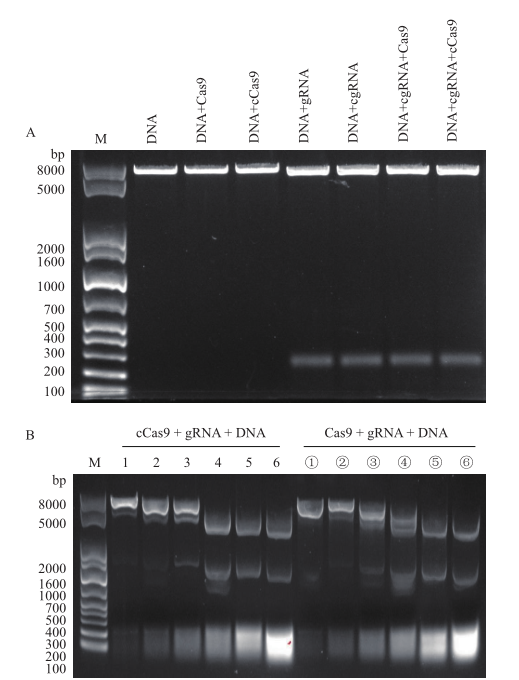
Fig.2 In vitro activity verification of RNP complex A: Negative control. B: Experimental group, 1-6(①-⑥)respectively indicates the addition amount of cCas9(Cas9),gRNA,and DNA at different concentrations. The addition amount of the three is shown in Table 3. M:DNA molecular mass marker

Fig.4 Verification of positive transformants by PCR M:DNA molecular mass marker;1-11 are different positive transformants;CK is the original strain as control
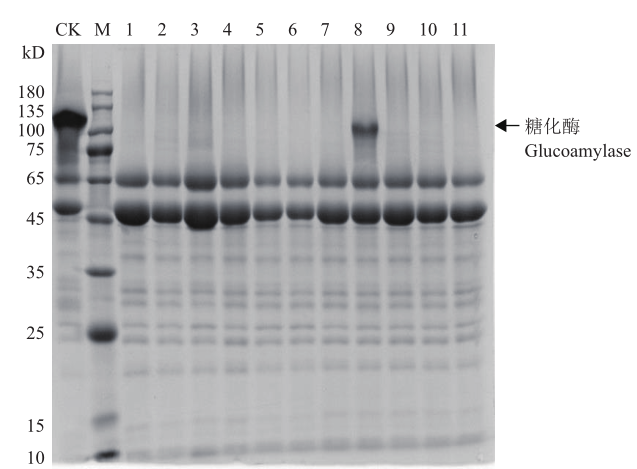
Fig.5 SDS-PAGE analysis of supernatant of transformant fermentation broth M:Protein molecular mass marker;1-11 are the fermentation broth supernatants of different transformants;CK is the original strain as control
| [1] |
Khadempar S, Familghadakchi S, Motlagh RA, et al. CRISPR-Cas9 in genome editing:Its function and medical applications[J]. J Cell Physiol, 2019, 234(5):5751-5761.
doi: 10.1002/jcp.27476 pmid: 30362544 |
| [2] | Chandrasekaran AP, Song M, Kim KS, et al. Different methods of delivering CRISPR/Cas 9 into cells[M]// Progress in Molecular Biology and Translational Science. Amsterdam:Elsevier, 2018:157-176. |
| [3] |
Song R, Zhai Q, Sun L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi:progress and perspective[J]. Appl Microbiol Biotechnol, 2019, 103(17):6919-6932.
doi: 10.1007/s00253-019-10007-w URL |
| [4] |
Krappmann S. CRISPR-Cas9, the new kid on the block of fungal molecular biology[J]. Med Mycol, 2017, 55(1):16-23.
pmid: 27811178 |
| [5] |
Salazar-Cerezo S, Kun RS, de Vries RP, et al. CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer[J]. Enzyme Microb Technol, 2020, 133:109463.
doi: 10.1016/j.enzmictec.2019.109463 URL |
| [6] |
Batool A, Malik F, Andrabi KI. Expansion of the CRISPR/cas genome-sculpting toolbox:innovations, applications and challenges[J]. Mol Diagn Ther, 2021, 25(1):41-57.
doi: 10.1007/s40291-020-00500-8 pmid: 33185860 |
| [7] |
Liu R, Chen L, Jiang YP, et al. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J]. Cell Discov, 2015, 1:15007.
doi: 10.1038/celldisc.2015.7 URL |
| [8] | 方凯伦, 杨辉. CRISPR/Cas工具的开发和应用[J]. 科学通报, 2020, 65(11):973-990. |
|
Fang KL, Yang H. Advances and applications of CRISPR/Cas toolbox[J]. Chin Sci Bull, 2020, 65(11):973-990.
doi: 10.1360/TB-2019-0806 URL |
|
| [9] |
Nødvig CS, Nielsen JB, Kogle ME, et al. A CRISPR-Cas9 system for genetic engineering of filamentous fungi[J]. PLoS One, 2015, 10(7):e0133085.
doi: 10.1371/journal.pone.0133085 URL |
| [10] |
Ullah M, Xia L, Xie SX, et al. CRISPR/Cas9-based genome engineering:a new breakthrough in the genetic manipulation of filamentous fungi[J]. Biotechnol Appl Biochem, 2020, 67(6):835-851.
doi: 10.1002/bab.2077 URL |
| [11] |
Zhang S, Shen JT, Li DL, et al. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing[J]. Theranostics, 2021, 11(2):614-648.
doi: 10.7150/thno.47007 pmid: 33391496 |
| [12] |
Mei YZ, Zhu YL, Huang PW, et al. Strategies for gene disruption and expression in filamentous fungi[J]. Appl Microbiol Biotechnol, 2019, 103(15):6041-6059.
doi: 10.1007/s00253-019-09953-2 URL |
| [13] | Wang Q, Coleman JJ. Progress and challenges:development and implementation of CRISPR/Cas9 technology in filamentous fungi[J]. Comput Struct Biotechnol J, 2019, 17:761-769. |
| [14] |
Wang R, Graham S, Gao L, et al. Editing the immune system in vivo in mice using CRISPR/Cas9 ribonucleoprotein(RNP)-mediated gene editing of transplanted hematopoietic stem cells[J]. Methods, 2021, 194:30-36.
doi: 10.1016/j.ymeth.2021.01.001 pmid: 33422676 |
| [15] |
Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells[J]. J Exp Med, 2018, 215(3):985-997.
doi: 10.1084/jem.20171626 URL |
| [16] |
Park J, Choe S. DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants[J]. Transgenic Res, 2019, 28(suppl 2):61-64.
doi: 10.1007/s11248-019-00136-3 URL |
| [17] |
Khatodia S, Bhatotia K, Tuteja N. Development of CRISPR/Cas9 mediated virus resistance in agriculturally important crops[J]. Bioengineered, 2017, 8(3):274-279.
doi: 10.1080/21655979.2017.1297347 URL |
| [18] |
Wang SX, Chen HQ, Tang X, et al. Molecular tools for gene manipulation in filamentous fungi[J]. Appl Microbiol Biotechnol, 2017, 101(22):8063-8075.
doi: 10.1007/s00253-017-8486-z URL |
| [19] |
Deng HX, Gao RJ, Liao XR, et al. CRISPR system in filamentous fungi:Current achievements and future directions[J]. Gene, 2017, 627:212-221.
doi: 10.1016/j.gene.2017.06.019 URL |
| [20] |
Kuivanen J, Korja V, Holmström S, et al. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes[J]. Fungal Biol Biotechnol, 2019, 6:3.
doi: 10.1186/s40694-019-0066-9 pmid: 30923622 |
| [21] |
Hao ZZ, Su XY. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex[J]. BMC Biotechnol, 2019, 19(1):2.
doi: 10.1186/s12896-018-0498-y URL |
| [22] |
Liang Z, Chen KL, Yan Y, et al. Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes[J]. Plant Biotechnol J, 2018, 16(12):2053-2062.
doi: 10.1111/pbi.12938 pmid: 29723918 |
| [23] |
Jackson RN, Wiedenheft B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses[J]. Mol Cell, 2015, 58(5):722-728.
doi: 10.1016/j.molcel.2015.05.023 pmid: 26028539 |
| [24] |
van Hartingsveldt W, Mattern IE, van Zeijl CM, et al. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene[J]. Mol Gen Genet, 1987, 206(1):71-75.
doi: 10.1007/BF00326538 URL |
| [25] |
Foster AJ, Martin-Urdiroz M, Yan X, et al. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus[J]. Sci Rep, 2018, 8(1):14355.
doi: 10.1038/s41598-018-32702-w URL |
| [26] | 潘海峰, 杨晗, 于思远, 等. 基于体外组装核糖核蛋白形式的CRISPR/Cas9基因编辑方法研究进展[J]. 中国生物工程杂志, 2019, 39(1):71-76. |
| Pan HF, Yang H, Yu SY, et al. Progress in gene editing methods of CRISPR/Cas9 based on in vitro assembly of ribonucleoprotein[J]. China Biotechnol, 2019, 39(1):71-76. | |
| [27] |
Zou G, Xiao ML, Chai SX, et al. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents[J]. Microb Biotechnol, 2021, 14(6):2343-2355.
doi: 10.1111/1751-7915.13652 URL |
| [28] |
Zhu SY, Xu Y, Yu XW. Improved homologous expression of the acidic lipase from Aspergillus niger[J]. J Microbiol Biotechnol, 2020, 30(2):196-205.
doi: 10.4014/jmb.1906.06028 URL |
| [29] |
Cairns TC, Nai C, Meyer V. How a fungus shapes biotechnology:100 years of Aspergillus niger research[J]. Fungal Biol Biotechnol, 2018, 5(1):1-14.
doi: 10.1186/s40694-018-0045-6 URL |
| [1] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [2] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [3] | TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates [J]. Biotechnology Bulletin, 2023, 39(11): 205-216. |
| [4] | LI Hai-li, LANG Li-min, ZHANG Qing-xian, YOU Yi, ZHU Wen-hao, WANG Zhi-fang, ZHANG Li-xian, WANG Ke-ling. Identification and Drug Resistance of Escherichia coli Simultaneously Producing Carbapenemase NDM-1 and NDM-5 [J]. Biotechnology Bulletin, 2022, 38(9): 106-115. |
| [5] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [6] | ZHAO Yan-kun, LIU Hui-min, MENG Lu, WANG Cheng, WANG Jia-qi, ZHENG Nan. Research Progress in Heteroresistance of Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(9): 59-71. |
| [7] | SUN Man-luan, GE Sai, BU Jia, ZHU Zhuang-yan. Regulation Mechanism of Ribonucleases in Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(3): 234-245. |
| [8] | REN Si-yu, CHENG Xin-kuan, ZHANG Yu-hui, ZHUANG Jian-wen, MA Long. Identification of Two New Types of L-threonine Aldolases and Development of Its Activity Detection Method [J]. Biotechnology Bulletin, 2021, 37(3): 233-240. |
| [9] | WANG Kai-kai, WANG Xiao-lu, SU Xiao-yun, ZHANG Jie. Optimization and Application of Double-plasmid CRISPR-Cas9 System in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(12): 252-264. |
| [10] | FENG Zhi-mei, ZHAO Ya-tong, LIU Ye-xue, LU Fu-ping, LI Yu. Recombinant Expression of NADPH-Dependent Mannitol Dehydrogenase and Transformation Conditions of Mannitol [J]. Biotechnology Bulletin, 2017, 33(8): 186-191. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||