








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (12): 252-264.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0618
Previous Articles Next Articles
WANG Kai-kai( ), WANG Xiao-lu, SU Xiao-yun(
), WANG Xiao-lu, SU Xiao-yun( ), ZHANG Jie(
), ZHANG Jie( )
)
Received:2021-05-11
Online:2021-12-26
Published:2022-01-19
Contact:
SU Xiao-yun,ZHANG Jie
E-mail:kkw_2012@163.com;suxiaoyun@caas.cn;zhangjie09@caas.cn
WANG Kai-kai, WANG Xiao-lu, SU Xiao-yun, ZHANG Jie. Optimization and Application of Double-plasmid CRISPR-Cas9 System in Escherichia coli[J]. Biotechnology Bulletin, 2021, 37(12): 252-264.
| 菌株Strain/质粒 Plasmid | 功能 Function | 来源 Source |
|---|---|---|
| 菌株Strain | ||
| 大肠杆菌Escherichia coli Top10 | 克隆宿主 | 天根 |
| MG1655(DE3) | 基本菌株 | 本研究 |
| MG1655-∆pfkA | 来源于MG1655(DE3),pfkA缺失突变株 | 本研究 |
| MG1655-∆pfkB | 来源于MG1655(DE3),pfkB缺失突变株 | 本研究 |
| MG1655-∆zwf | 来源于MG1655(DE3),zwf缺失突变株 | 本研究 |
| MG1655-∆ABE∷glpK | 来源于MG1655(DE3),nagABE基因位点替换为glpK的突变株 | 本研究 |
| 质粒 Plasmid | ||
| pRed_cas9_recA_poxb300 | Cas9供体质粒 | MolecularCloud |
| p∆pfkA | pfkA同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆pfkA | pfkA敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| p∆pfkB | pfkB同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆pfkB | pfkB敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| p∆zwf | zwf同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆zwf | zwf敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| p∆ABE∷glpK | nagABE同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆ABE∷glpK | nagABE敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
Table1 Strains and plasmids used in this study
| 菌株Strain/质粒 Plasmid | 功能 Function | 来源 Source |
|---|---|---|
| 菌株Strain | ||
| 大肠杆菌Escherichia coli Top10 | 克隆宿主 | 天根 |
| MG1655(DE3) | 基本菌株 | 本研究 |
| MG1655-∆pfkA | 来源于MG1655(DE3),pfkA缺失突变株 | 本研究 |
| MG1655-∆pfkB | 来源于MG1655(DE3),pfkB缺失突变株 | 本研究 |
| MG1655-∆zwf | 来源于MG1655(DE3),zwf缺失突变株 | 本研究 |
| MG1655-∆ABE∷glpK | 来源于MG1655(DE3),nagABE基因位点替换为glpK的突变株 | 本研究 |
| 质粒 Plasmid | ||
| pRed_cas9_recA_poxb300 | Cas9供体质粒 | MolecularCloud |
| p∆pfkA | pfkA同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆pfkA | pfkA敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| p∆pfkB | pfkB同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆pfkB | pfkB敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| p∆zwf | zwf同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆zwf | zwf敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| p∆ABE∷glpK | nagABE同源臂-gRNA供体质粒,用于双质粒CRISPR-Cas9系统 | 本研究 |
| pRA∆ABE∷glpK | nagABE敲除质粒,用于单质粒CRISPR-Cas9系统 | 本研究 |
| 说明Instruction | 引物Primer | 寡核苷酸序列 Oligonucleotide sequence(5'-3') |
|---|---|---|
| 双质粒pfkA基因敲除pfkA gene knockout(double plasmid system) | ||
| gRNA扩增 gRNA amplification | pfkA-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCGTGTCTGACAT-GATCAACCGGTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | pfkA-N-F | AATCACTAGTGAATTCGCGGCCGTCGGCATCTATATTTTATATAGCG |
| pfkA-N-R | GAAATCAGACTACCTCTGAACTTTGGAATGCAAAATGAAATCTGTTGC | |
| 下游同源臂扩增Downstream homology arm amplification | pfkA-C-F | TCCAAAGTTCAGAGGTAGTCTGATTTCGGAAAAAGGCAGATTC |
| pfkA-C-R | ATGCATCCAACGCGTTGGGAGCTCTCCCACCGTGTGACTGACGAATC | |
| 单质粒pfkA基因敲除pfkA gene knockout(single plasmid system) | ||
| pfkA-F1 | GATCTGTGTCTGACATGATCAACCGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAG | |
| pfkA-R1 | TATATAAAATATAGATGCCGAGGTCCCGCTTTGTTACAGAATGCTT | |
| pfkA-up-F1 | CGGGACCTCGGCATCTATATTTTATATAGCGCGTTACGCATGG | |
| pfkA-down-R1 | CGTTTTTGTCATGGCTTTCACCGTGTGACTGACG | |
| pfkA-F2 | TGAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTAT | |
| pfkA-R2 | CGGTTGATCATGTCAGACACAGATCTGACTCCATAACAGAGTACTCGC | |
| 双质粒pfkB基因敲除pfkB gene knockout(double plasmid system) | ||
| gRNA扩增 gRNA amplification | pfkB-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCACGTACATGTGGAAGCAAG-GTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | pfkB-N-F | CAATCACTAGTGAATTCGCGGCCGCAGCGACCAGGCAGTGGTGTGTC |
| pfkB-N-R | GGGGAATGTTTTTGCATTTCCTCCTATAGGCTGA | |
| 下游同源臂扩增Downstream homology arm amplification | pfkB-C-F | GGAGGAAATGCAAAAACATTCCCCCAGCATTGGGGGAATCATCAC |
| pfkB-C-R | TGCATCCAACGCGTTGGGAGCTCTCCTCAAAAGACGACTGATTGCCTGC | |
| 单质粒pfkB基因敲除pfkB gene knockout(single plasmid system) | ||
| pfkB-F1 | AGATCTCACGTACATGTGGAAGCAAGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAG | |
| pfkB-R1 | ACACCACTGCCTGGTCGCTGGGTCCCGCTTTGTTACAGAATGCTTTTAAT | |
| pfkB-up-F1 | AAAGCGGGACCCAGCGACCAGGCAGTGGTGTGTCCATACCAGGTCAT | |
| pfkB-down-R1 | GCGTTTTTGTCATGGCTTTTCAAAAGACGACTGATTGCCTGCCACAT | |
| pfkB-F2 | TTGAAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTATAATCA | |
| pfkB-R2 | CTTGCTTCCACATGTACGTGAGATCTGACTCCATAACAGAGTACTCG | |
| 双质粒zwf基因敲除zwf gene knockout(double plasmid system) | ||
| gRNA扩增 gRNA amplification | zwf-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCGCGTGCTGACTGGGATA- AAGGTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | zwf-N-F1 | AATCACTAGTGAATTCGCGGCCGCGTGTCCATGCTGCGACAGAAACG |
| zwf-N-R1 | GCAGATAGTCATTCTCCTTAAGTTAACTAACCCGGTACTTAAGC | |
| 下游同源臂扩增Downstream homology arm amplification | zwf-C-F1 | AGTTAACTTAAGGAGAATGACTATCTGCGCTTATCCTTTATGG |
| zwf-C-R1 | TCCAACGCGTTGGGAGCTCTCCCGCATTCGCTTCATGCAGGGCTTTAC | |
| 单质粒zwf基因敲除zwf gene knockout(single plasmid system) | ||
| zwf-F1 | ATGGAGTCAGATCTGCGTGCTGACTGGGATAAAGGTTTTAG- AGCTAGAAATAGCAAGTTAAAATAAGG | |
| zwf-R1 | TCTGTCGCAGCATGGACACGGGTCCCGCTTTGTTACAGAATGCT | |
| zwf -up-F1 | AAGCGGGACCCGTGTCCATGCTGCGACAGAAACGATTCACCGTCG | |
| zwf -down-R1 | GTTTTTGTCATGGCTTTCGCATTCGCTTCATGCAGGGCTTTACGAAT | |
| zwf -F2 | ATGAAGCGAATGCGAAAGCCATGACAAAAACGCGTAACAAAAGTG | |
| zwf -R2 | CTTTATCCCAGTCAGCACGCAGATCTGACTCCATAACAGAGTACTCGCCTAT | |
| 双质粒glpK基因与nagABE替换nagABE replacement by glpK gene(double plasmid system) | ||
| gRNA扩增 gRNA amplification | nagABE-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCCTGCAGCGCCAG- TGCTTTCGTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | nagABE-N-F | AATCACTAGTGAATTCGCGGCCGAGTTTTCTACCTGAATATGCGGC |
| nagABE-N-R | CTTATTGAGGTGAATAATGACACCAGGCGGACAAGCTCAGATAGG | |
| 下游同源臂扩增Downstream homology arm amplification | nagABE-C-F | TTGGTGACAAAACTCACAATCTGCTTTATGCCTGATGCGACGC |
| nagABE-C-R | TGCATCCAACGCGTTGGGAGCTCTCCGGAGCTGATCAAAATAATCGG | |
| nagB启动子扩增nagB promotor amplification | nagB-Pm-F | TGAGCTTGTCCGCCTGGTGTCATTATTCACCTCAATAAGTAAAATGTA |
| nagB-Pm-R | CAAGCGTCGCATCAGGCATAAAGCAGATTGTGAGTTTTGTCACC | |
| T7启动子扩增 T7 promotor amplification | nagABE-T7-F | TTTGGTGACAAAACTCACAAGGATCGAGATCTCGATCCCGCGAA |
| nagABE-T7-R | GCAACGATATATTTTTTTTCAGTCATGGTATATCTCCTTCTTAAAGTT | |
| glpK基因扩增 glpK gene amplification | pichi-glpk-F | CTTTAAGAAGGAGATATACCATGGGAAAAGACTATACACCAC |
| pichi-glpk-R | CGTCGCATCAGGCATAAAGCAGATTAAGCAGTGTCCTTAAGCCAGCCC | |
| 单质粒glpK基因整合glpK gene integration(single plasmid system) | ||
| ABE-F1 | GTCAGATCTCCTGCAGCGCCAGTGCTTTCGTTTTAGAGCTAGAAATAGCAAGTTAAAA- TAAGG | |
| ABE -R1 | GCATATTCAGGTAGAAAACTGGTCCCGCTTTGTTACAGAATGC | |
| ABE-up-F1 | GCGGGACCAGTTTTCTACCTGAATATGCGGCATGTAATGAATTTTGCCGCTGTC | |
| ABE-down-R1 | TGTTACGCGTTTTTGTCATGGCTTTGGAGCTGATCAAAATAATCGGAG | |
| ABE-F2 | CTCCAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTATAATCACGGC | |
| ABE-R2 | CGAAAGCACTGGCGCTGCAGGAGATCTGACTCCATAACAGAGTACTC | |
| 突变株检测引物Detection primer for mutants | ||
| zwf-JC-F | ACGGCACAAACACCGCAGGC | |
| zwf-JC-R | ACATGATCAAGCGTTGCCATTGC | |
| A-JC-F | TTATCCTTGTCTCGTTTCAGCGTTGGGTGGTGCG | |
| A-JC-R | GTTTGATCCACTCTTTATCAATC | |
| B-JC-F | CATCGCGCTCTCGATAGCCGTTAT | |
| B-JC-R | GCGCAAATGCCATGCGGCATGGA | |
| ABE-JC-R | TTCAACGCTGCGGTCGCGGTAC | |
| ABE-JC-F | GGTTCCGCGATGGACACGCACC | |
Table 2 Primers used in this study
| 说明Instruction | 引物Primer | 寡核苷酸序列 Oligonucleotide sequence(5'-3') |
|---|---|---|
| 双质粒pfkA基因敲除pfkA gene knockout(double plasmid system) | ||
| gRNA扩增 gRNA amplification | pfkA-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCGTGTCTGACAT-GATCAACCGGTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | pfkA-N-F | AATCACTAGTGAATTCGCGGCCGTCGGCATCTATATTTTATATAGCG |
| pfkA-N-R | GAAATCAGACTACCTCTGAACTTTGGAATGCAAAATGAAATCTGTTGC | |
| 下游同源臂扩增Downstream homology arm amplification | pfkA-C-F | TCCAAAGTTCAGAGGTAGTCTGATTTCGGAAAAAGGCAGATTC |
| pfkA-C-R | ATGCATCCAACGCGTTGGGAGCTCTCCCACCGTGTGACTGACGAATC | |
| 单质粒pfkA基因敲除pfkA gene knockout(single plasmid system) | ||
| pfkA-F1 | GATCTGTGTCTGACATGATCAACCGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAG | |
| pfkA-R1 | TATATAAAATATAGATGCCGAGGTCCCGCTTTGTTACAGAATGCTT | |
| pfkA-up-F1 | CGGGACCTCGGCATCTATATTTTATATAGCGCGTTACGCATGG | |
| pfkA-down-R1 | CGTTTTTGTCATGGCTTTCACCGTGTGACTGACG | |
| pfkA-F2 | TGAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTAT | |
| pfkA-R2 | CGGTTGATCATGTCAGACACAGATCTGACTCCATAACAGAGTACTCGC | |
| 双质粒pfkB基因敲除pfkB gene knockout(double plasmid system) | ||
| gRNA扩增 gRNA amplification | pfkB-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCACGTACATGTGGAAGCAAG-GTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | pfkB-N-F | CAATCACTAGTGAATTCGCGGCCGCAGCGACCAGGCAGTGGTGTGTC |
| pfkB-N-R | GGGGAATGTTTTTGCATTTCCTCCTATAGGCTGA | |
| 下游同源臂扩增Downstream homology arm amplification | pfkB-C-F | GGAGGAAATGCAAAAACATTCCCCCAGCATTGGGGGAATCATCAC |
| pfkB-C-R | TGCATCCAACGCGTTGGGAGCTCTCCTCAAAAGACGACTGATTGCCTGC | |
| 单质粒pfkB基因敲除pfkB gene knockout(single plasmid system) | ||
| pfkB-F1 | AGATCTCACGTACATGTGGAAGCAAGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAG | |
| pfkB-R1 | ACACCACTGCCTGGTCGCTGGGTCCCGCTTTGTTACAGAATGCTTTTAAT | |
| pfkB-up-F1 | AAAGCGGGACCCAGCGACCAGGCAGTGGTGTGTCCATACCAGGTCAT | |
| pfkB-down-R1 | GCGTTTTTGTCATGGCTTTTCAAAAGACGACTGATTGCCTGCCACAT | |
| pfkB-F2 | TTGAAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTATAATCA | |
| pfkB-R2 | CTTGCTTCCACATGTACGTGAGATCTGACTCCATAACAGAGTACTCG | |
| 双质粒zwf基因敲除zwf gene knockout(double plasmid system) | ||
| gRNA扩增 gRNA amplification | zwf-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCGCGTGCTGACTGGGATA- AAGGTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | zwf-N-F1 | AATCACTAGTGAATTCGCGGCCGCGTGTCCATGCTGCGACAGAAACG |
| zwf-N-R1 | GCAGATAGTCATTCTCCTTAAGTTAACTAACCCGGTACTTAAGC | |
| 下游同源臂扩增Downstream homology arm amplification | zwf-C-F1 | AGTTAACTTAAGGAGAATGACTATCTGCGCTTATCCTTTATGG |
| zwf-C-R1 | TCCAACGCGTTGGGAGCTCTCCCGCATTCGCTTCATGCAGGGCTTTAC | |
| 单质粒zwf基因敲除zwf gene knockout(single plasmid system) | ||
| zwf-F1 | ATGGAGTCAGATCTGCGTGCTGACTGGGATAAAGGTTTTAG- AGCTAGAAATAGCAAGTTAAAATAAGG | |
| zwf-R1 | TCTGTCGCAGCATGGACACGGGTCCCGCTTTGTTACAGAATGCT | |
| zwf -up-F1 | AAGCGGGACCCGTGTCCATGCTGCGACAGAAACGATTCACCGTCG | |
| zwf -down-R1 | GTTTTTGTCATGGCTTTCGCATTCGCTTCATGCAGGGCTTTACGAAT | |
| zwf -F2 | ATGAAGCGAATGCGAAAGCCATGACAAAAACGCGTAACAAAAGTG | |
| zwf -R2 | CTTTATCCCAGTCAGCACGCAGATCTGACTCCATAACAGAGTACTCGCCTAT | |
| 双质粒glpK基因与nagABE替换nagABE replacement by glpK gene(double plasmid system) | ||
| gRNA扩增 gRNA amplification | nagABE-gRNA-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCCTGCAGCGCCAG- TGCTTTCGTTTTAGAGCTAGAAATAGCAAG |
| gRNA-R | TCGGATCCACTAGTAACCATC | |
| 上游同源臂扩增Upstream homology arm amplification | nagABE-N-F | AATCACTAGTGAATTCGCGGCCGAGTTTTCTACCTGAATATGCGGC |
| nagABE-N-R | CTTATTGAGGTGAATAATGACACCAGGCGGACAAGCTCAGATAGG | |
| 下游同源臂扩增Downstream homology arm amplification | nagABE-C-F | TTGGTGACAAAACTCACAATCTGCTTTATGCCTGATGCGACGC |
| nagABE-C-R | TGCATCCAACGCGTTGGGAGCTCTCCGGAGCTGATCAAAATAATCGG | |
| nagB启动子扩增nagB promotor amplification | nagB-Pm-F | TGAGCTTGTCCGCCTGGTGTCATTATTCACCTCAATAAGTAAAATGTA |
| nagB-Pm-R | CAAGCGTCGCATCAGGCATAAAGCAGATTGTGAGTTTTGTCACC | |
| T7启动子扩增 T7 promotor amplification | nagABE-T7-F | TTTGGTGACAAAACTCACAAGGATCGAGATCTCGATCCCGCGAA |
| nagABE-T7-R | GCAACGATATATTTTTTTTCAGTCATGGTATATCTCCTTCTTAAAGTT | |
| glpK基因扩增 glpK gene amplification | pichi-glpk-F | CTTTAAGAAGGAGATATACCATGGGAAAAGACTATACACCAC |
| pichi-glpk-R | CGTCGCATCAGGCATAAAGCAGATTAAGCAGTGTCCTTAAGCCAGCCC | |
| 单质粒glpK基因整合glpK gene integration(single plasmid system) | ||
| ABE-F1 | GTCAGATCTCCTGCAGCGCCAGTGCTTTCGTTTTAGAGCTAGAAATAGCAAGTTAAAA- TAAGG | |
| ABE -R1 | GCATATTCAGGTAGAAAACTGGTCCCGCTTTGTTACAGAATGC | |
| ABE-up-F1 | GCGGGACCAGTTTTCTACCTGAATATGCGGCATGTAATGAATTTTGCCGCTGTC | |
| ABE-down-R1 | TGTTACGCGTTTTTGTCATGGCTTTGGAGCTGATCAAAATAATCGGAG | |
| ABE-F2 | CTCCAAAGCCATGACAAAAACGCGTAACAAAAGTGTCTATAATCACGGC | |
| ABE-R2 | CGAAAGCACTGGCGCTGCAGGAGATCTGACTCCATAACAGAGTACTC | |
| 突变株检测引物Detection primer for mutants | ||
| zwf-JC-F | ACGGCACAAACACCGCAGGC | |
| zwf-JC-R | ACATGATCAAGCGTTGCCATTGC | |
| A-JC-F | TTATCCTTGTCTCGTTTCAGCGTTGGGTGGTGCG | |
| A-JC-R | GTTTGATCCACTCTTTATCAATC | |
| B-JC-F | CATCGCGCTCTCGATAGCCGTTAT | |
| B-JC-R | GCGCAAATGCCATGCGGCATGGA | |
| ABE-JC-R | TTCAACGCTGCGGTCGCGGTAC | |
| ABE-JC-F | GGTTCCGCGATGGACACGCACC | |
| 基因编辑位点 Gene editing site | 单质粒CRISPR-Cas9系统编辑效率 Gene editing efficiency of the single-plasmid CRISPR-Cas9 system/% | 双质粒CRISPR-Cas9系统编辑效率 Gene editing efficiency of the double-plasmid CRISPR-Cas9 system/% |
|---|---|---|
| pfkA | 6 | 37 |
| pfkB | 0 | 6 |
| zwf | 37 | 100 |
| nagABE(glpK Integration) | 0 | 10 |
Table 3 Comparison of gene editing efficiency
| 基因编辑位点 Gene editing site | 单质粒CRISPR-Cas9系统编辑效率 Gene editing efficiency of the single-plasmid CRISPR-Cas9 system/% | 双质粒CRISPR-Cas9系统编辑效率 Gene editing efficiency of the double-plasmid CRISPR-Cas9 system/% |
|---|---|---|
| pfkA | 6 | 37 |
| pfkB | 0 | 6 |
| zwf | 37 | 100 |
| nagABE(glpK Integration) | 0 | 10 |
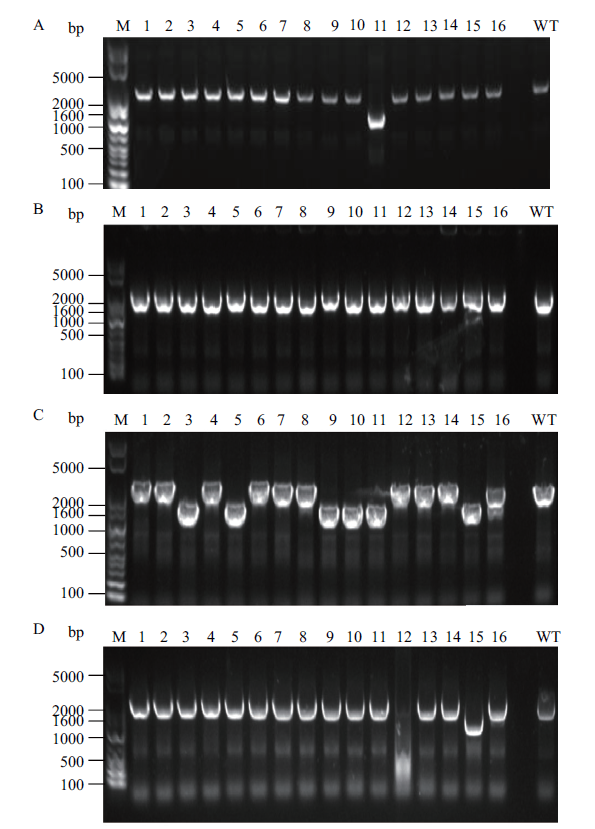
Fig. 2 Identification of pfkA and pfkB deletion by gel electrophoresis A:pfkA knockout using single-plasmid CRISPR-Cas9 system. B:pfkB knockout using single-plasmid CRISPR-Cas9 system. C:pfkA knockout using double-plasmid CRISPR-Cas9 system. D:pfkB knockout using double-plasmid CRISPR-Cas9 system. The target band size of the pfkA deleted strain is 1 327 bp,and the related band size of the wild-type strain is 2 290 bp. The target band size of the pfkB deleted strain is 1 287 bp,and the related band size of wild-type strain is 2 217 bp
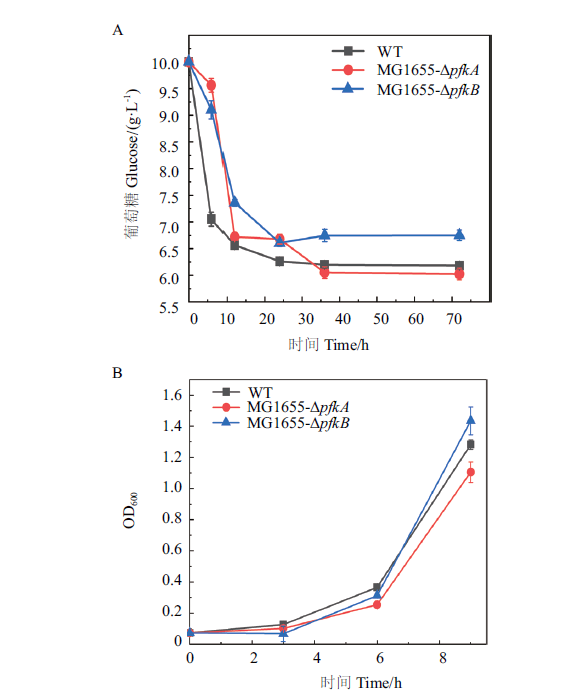
Fig. 3 Verification of the phenotypes of pfkA and pfkB deleted mutants A:Effect of pfkA or pfkB knockout on glucose metabolism. B:Effect of pfkA or pfkB knockout on cell growth

Fig. 4 Identification of zwf deletion by gel electrophoresis A:zwf knockout using single-plasmid CRISPR-Cas9 system. B:zwf knockout using double-plasmid CRISPR-Cas9 system. The target band size of the zwf deleted strain is 1 429 bp,and the related band size of the wild-type strain is 2 905 bp
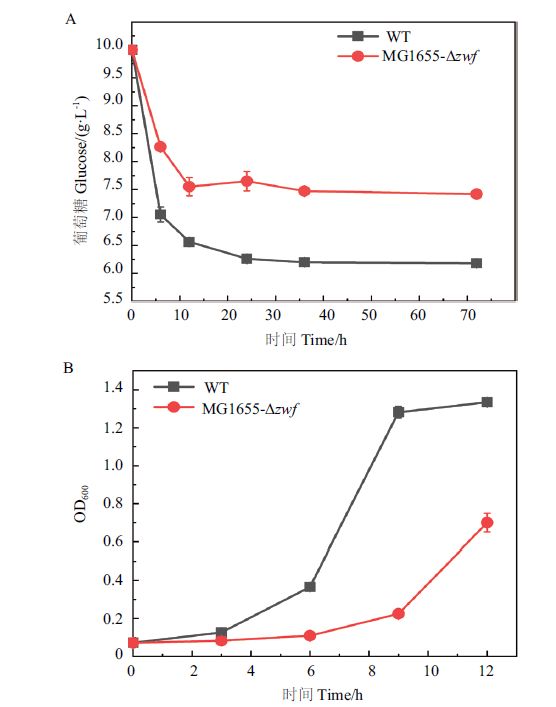
Fig. 5 Characterization of the phenotypes of zwf deleted mutant A:Effect of zwf knockout on glucose metabolism. B:Effect of zwf knockout on cell growth
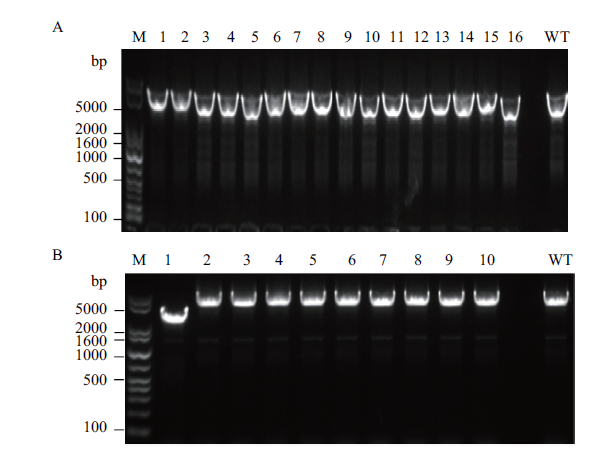
Fig. 6 Identification of nagABE replaced by glpK via gel electrophoresis A:nagABE replaced by glpK gene using single-plasmid CRISPR-Cas9 system. B:nagABE replaced by glpK gene using double-plasmid CRISPR-Cas9 system. The target band size of the glpK integrated strain is 3 317 bp,and the related band size of the wild-type strain is 5 626 bp
| [1] | Hatada I, Horii T. CRISPR/Cas9[J]. Methods Mol Biol, 2017, 1630:37-42. |
| [2] |
Xing HL, Dong L, Wang ZP, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants[J]. BMC Plant Biology, 2014, 14(1):327.
doi: 10.1186/s12870-014-0327-y URL |
| [3] |
Li W, Teng F, Li T, et al. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems[J]. Nat Biotechnol, 2013, 31(8):684-686.
doi: 10.1038/nbt.2652 URL |
| [4] |
Weninger A, Hatzl AM, Schmid C, et al. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris[J]. J Biotechnol, 2016, 235:139-149.
doi: 10.1016/j.jbiotec.2016.03.027 pmid: 27015975 |
| [5] |
Zhao D, Yuan S, Xiong B, et al. Development of a fast and easy method for Escherichia coli genome editing with CRISPR/Cas9[J]. Microb Cell Factories, 2016, 15(1):205.
doi: 10.1186/s12934-016-0605-5 URL |
| [6] |
Zhang JH, Adikaram P, Pandey M, et al. Optimization of genome editing through CRISPR-Cas9 engineering[J]. Bioengineered, 2016, 7(3):166-174.
doi: 10.1080/21655979.2016.1189039 URL |
| [7] | Hryhorowicz M, Lipiński D, Zeyland J, et al. CRISPR/Cas9 immune system as a tool for genome engineering[J]. Arch Immunol Ther Exp(Warsz), 2017, 65(3):233-240. |
| [8] |
Chung ME, Yeh IH, Sung LY, et al. Enhanced integration of large DNA into E. coli chromosome by CRISPR/Cas9[J]. Biotechnol Bioeng, 2017, 114(1):172-183.
doi: 10.1002/bit.v114.1 URL |
| [9] |
Hatti-Kaul R, Chen L, Dishisha T, et al. Lactic acid bacteria:from starter cultures to producers of chemicals[J]. FEMS Microbiol Lett, 2018, 365(20). DOI: 10.1093/femsle/fny213.
doi: 10.1093/femsle/fny213 |
| [10] |
Gleizer S, Ben R, Bar YM, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2[J]. Cell, 2019, 179(6):1255-1263.e1212.
doi: S0092-8674(19)31230-9 pmid: 31778652 |
| [11] |
Zhang S, Guo F, Yan W, et al. Recent advances of CRISPR/Cas9-based genetic engineering and transcriptional regulation in industrial biology[J]. Front Bioeng Biotechnol, 2020, 7:459-459.
doi: 10.3389/fbioe.2019.00459 URL |
| [12] |
Tovilla-Coutiño DB, Momany C, Eiteman MA. Engineered citrate synthase alters acetate accumulation in Escherichia coli[J]. Metab Eng, 2020, 61:171-180.
doi: S1096-7176(20)30105-1 pmid: 32569710 |
| [13] |
Heo MJ, Jung HM, Um J, et al. Controlling citrate synthase expression by CRISPR/Cas9 genome editing for N-butanol production in Escherichia coli[J]. ACS Synthetic Biology, 2017, 6(2):182-189.
doi: 10.1021/acssynbio.6b00134 URL |
| [14] |
Ferguson GP, Nikolaev Y, Mclaggan D, et al. Survival during exposure to the electrophilic reagent N-ethylmaleimide in Escherichia coli:role of KefB and KefC potassium channels[J]. J Bacteriol, 1997, 179(4):1007-1012.
pmid: 9023177 |
| [15] |
Jiang Y, Chen B, Duan C, et al. Erratum for Jiang et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system[J]. Appl Environ Microbiol, 2016, 82(12):3693.
doi: 10.1128/AEM.01181-16 URL |
| [16] |
Lovingshimer MR, Siegele D, Reinhart GD. Construction of an inducible, pfkA and pfkB deficient strain of Escherichia coli for the expression and purification of phosphofructokinase from bacterial sources[J]. Protein Expr Purif, 2006, 46(2):475-482.
doi: 10.1016/j.pep.2005.09.015 URL |
| [17] |
Woo JE, Seong HJ, Lee SY, et al. Metabolic engineering of Escherichia coli for the production of hyaluronic acid from glucose and galactose[J]. Front Bioeng Biotechnol, 2019, 7:351-351.
doi: 10.3389/fbioe.2019.00351 URL |
| [18] |
Tokuyama K, Ohno S, Yoshikawa K, et al. Increased 3-hydroxypropionic acid production from glycerol, by modification of central metabolism in Escherichia coli[J]. Microb Cell Fact, 2014, 13:64.
doi: 10.1186/1475-2859-13-64 pmid: 24885133 |
| [19] |
Pettigrew DW, Ma DP, Conrad CA, et al. Escherichia coli glycerol kinase. Cloning and sequencing of the glpK gene and the primary structure of the enzyme[J]. J Biol Chem, 1988, 263(1):135-139.
pmid: 2826434 |
| [20] |
Yoshida N, Sato M. Plasmid uptake by bacteria:a comparison of methods and efficiencies[J]. Appl Microbiol Biotechnol, 2009, 83(5):791-798.
doi: 10.1007/s00253-009-2042-4 pmid: 19471921 |
| [21] |
Zhang J, Zong W, Hong W, et al. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production[J]. Metab Eng, 2018, 47:49-59.
doi: S1096-7176(18)30059-4 pmid: 29530750 |
| [22] |
Thompson MG, Sedaghatian N, Barajas JF, et al. Isolation and characterization of novel mutations in the pSC101 origin that increase copy number[J]. Sci. Rep., 2018, 8(1):1590.
doi: 10.1038/s41598-018-20016-w URL |
| [23] | Karcher SJ. Recombinant DNA cloning[M]//Karcher SJ. Molecular Biology. San Diego: Academic Press, 1995:45-134. |
| [24] |
Larson MH, Gilbert LA, Wang X, et al. CRISPR interference(CRISPRi)for sequence-specific control of gene expression[J]. Nat Protoc, 2013, 8(11):2180-2196.
doi: 10.1038/nprot.2013.132 URL |
| [25] |
Moradpour M, Abdulah SNA. CRISPR/dCas9 platforms in plants:strategies and applications beyond genome editing[J]. Plant Biotechnol J, 2020, 18(1):32-44.
doi: 10.1111/pbi.13232 pmid: 31392820 |
| [26] |
Bao A, Burritt DJ, Chen H, et al. The CRISPR/Cas9 system and its applications in crop genome editing[J]. Crit Rev Biotechnol, 2019, 39(3):321-336.
doi: 10.1080/07388551.2018.1554621 URL |
| [27] |
Gupta D, Bhattacharjee O, Mandal D, et al. CRISPR-Cas9 system:a new-fangled dawn in gene editing[J]. Life Sci, 2019, 232:116636.
doi: 10.1016/j.lfs.2019.116636 URL |
| [28] |
Zhan T, Rindtorff N, Betge J, et al. CRISPR/Cas9 for cancer research and therapy[J]. Semin Cancer Biol, 2019, 55:106-119.
doi: 10.1016/j.semcancer.2018.04.001 URL |
| [29] | 武林琳, 竹梦婕, 王咪, 等. CRISPR/Cas9技术在农作物中应用的局限及改进[J]. 现代农业科技, 2020(22):26-29. |
| Wu LL, Zhu MJ, Wang M, et al. The limitation and improvement of CRISPR/Cas9 technology in crops application[J]. Modern Agricultural Science and Technology, 2020(22):26-29. | |
| [30] | 李秋月, 王小柯, 张亚飞, 等. 通过优化gRNA设计提高CRISPR/Cas9系统中基因编辑效率的方法[J]. 分子植物育种, 2019, 17(10):3274-3282. |
| Li QY, Wang XK, Zhang YF, et al. Methods to improve the efficiency of gene editing in CRISPR/Cas9 system by optimizing gRNA design[J]. Molecular Plant Breeding, 17(10):3274-3282. | |
| [31] |
Baker O, Tsurkan S, Fu J, et al. The contribution of homology arms to nuclease-assisted genome engineering[J]. Nucleic Acids Res, 2017, 45(13):8105-8115.
doi: 10.1093/nar/gkx497 URL |
| [32] |
Su B, Song D, Zhu H. Homology-dependent recombination of large synthetic pathways into E. coli genome via λ-Red and CRISPR/Cas9 dependent selection methodology[J]. Microb Cell Factories, 2020, 19(1):108.
doi: 10.1186/s12934-020-01360-x URL |
| [33] |
Fricke PM, Link T, Gätgens J, et al. A tunable L-arabinose-inducible expression plasmid for the acetic acid bacterium Gluconobacter oxydans[J]. Appl Microbiol Biotechnol, 2020, 104(21):9267-9282.
doi: 10.1007/s00253-020-10905-4 URL |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [3] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [4] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [5] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [6] | TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates [J]. Biotechnology Bulletin, 2023, 39(11): 205-216. |
| [7] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [8] | LI Hai-li, LANG Li-min, ZHANG Qing-xian, YOU Yi, ZHU Wen-hao, WANG Zhi-fang, ZHANG Li-xian, WANG Ke-ling. Identification and Drug Resistance of Escherichia coli Simultaneously Producing Carbapenemase NDM-1 and NDM-5 [J]. Biotechnology Bulletin, 2022, 38(9): 106-115. |
| [9] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [10] | ZHAO Yan-kun, LIU Hui-min, MENG Lu, WANG Cheng, WANG Jia-qi, ZHENG Nan. Research Progress in Heteroresistance of Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(9): 59-71. |
| [11] | GAO Wei-xin, HUANG Huo-qing, ZHAO Jing, ZHANG Xin, YANG Ning, YANG Hao-meng. Construction and Activity Verification of Ribonucleoprotein Complex for Gene Editing [J]. Biotechnology Bulletin, 2022, 38(8): 60-68. |
| [12] | LAI Xin-tong, WANG Ke-lan, YOU Yu-xin, TAN Jun-jie. Recent Advances in CRISPR/Cas-based DNA Base Editing [J]. Biotechnology Bulletin, 2022, 38(6): 1-12. |
| [13] | ZHANG Hao, LI Zhe, GUO Kai, HUANG Yan-hua, HAO Yong-ren. Functional Analysis of TvGCN5 Gene Encoding Histone Acetylase from Trichoderma viride Tv-1511 [J]. Biotechnology Bulletin, 2022, 38(5): 136-148. |
| [14] | CHEN Ying-dan, ZHANG Yang, XIA Qiang, SUN Hong-xia. Gene Editing Technology of CRISPR/Cas and Its Applications in Microalgae Research [J]. Biotechnology Bulletin, 2022, 38(5): 257-268. |
| [15] | SUN Man-luan, GE Sai, BU Jia, ZHU Zhuang-yan. Regulation Mechanism of Ribonucleases in Escherichia coli [J]. Biotechnology Bulletin, 2022, 38(3): 234-245. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||