








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (8): 77-83.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1368
Previous Articles Next Articles
CAI Jia1,2( ), LIANG Zhen-yu1, HUANG Yu1, LU Yi-shan1, SHI gang1, JIAN Ji-chang1
), LIANG Zhen-yu1, HUANG Yu1, LU Yi-shan1, SHI gang1, JIAN Ji-chang1
Received:2021-10-29
Online:2022-08-26
Published:2022-09-14
Contact:
CAI Jia
E-mail:matrix924@foxmail.com
CAI Jia, LIANG Zhen-yu, HUANG Yu, LU Yi-shan, SHI gang, JIAN Ji-chang. Screening and Identifing the Interacting Proteins of Grouper(Epinephelus coioides)EcBAG3 Using Yeast Two-hybrid System[J]. Biotechnology Bulletin, 2022, 38(8): 77-83.
Fig. 2 Agarose gel electrophoresis for the detection of partial inserted cDNA fragments in library M:DL2000 DNA marker. 1-24:PCR products of inserted cDNA fragments
| 候选蛋白 Candidate protein | NCBI登录号 Accession number of NCBI |
|---|---|
| Heat Shock Cognate 71 kD Protein(Hsc71) | XP_033491860.1 |
| Heat Shock Protein 70(Hsp70) | AWD76391.1 |
| Filamin-A | XP_033484911.1 |
| Collagen,Type V,Alpha 2a | XP_033473099.1 |
| Zinc Finger Protein 217(ZNF217) | XP_033484552.1 |
| CCN Family Member 1(CCN1) | XP_033476535.1 |
| Suppressor Of Cytokine Signaling 9(SOCS9) | XP_033486547.1 |
Table 1 Alignments of positive clones screened through yeast two hybrid
| 候选蛋白 Candidate protein | NCBI登录号 Accession number of NCBI |
|---|---|
| Heat Shock Cognate 71 kD Protein(Hsc71) | XP_033491860.1 |
| Heat Shock Protein 70(Hsp70) | AWD76391.1 |
| Filamin-A | XP_033484911.1 |
| Collagen,Type V,Alpha 2a | XP_033473099.1 |
| Zinc Finger Protein 217(ZNF217) | XP_033484552.1 |
| CCN Family Member 1(CCN1) | XP_033476535.1 |
| Suppressor Of Cytokine Signaling 9(SOCS9) | XP_033486547.1 |
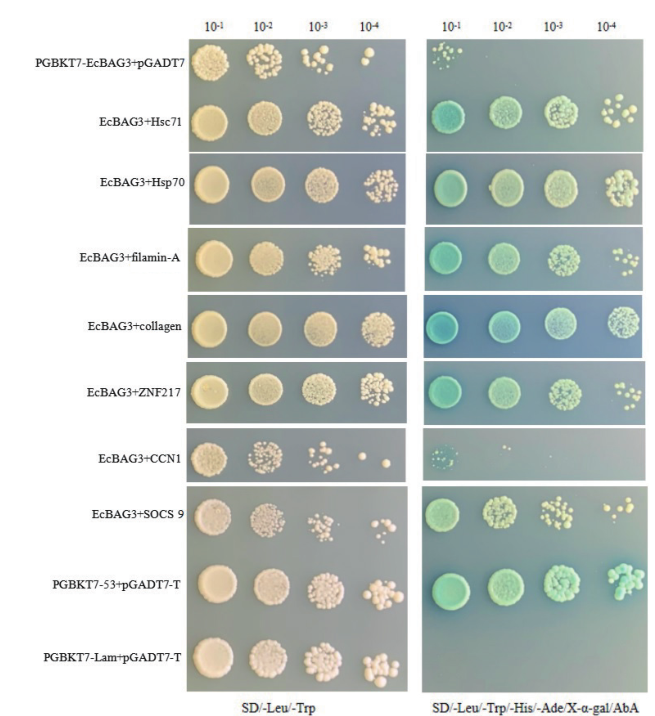
Fig. 5 Verification of EcBAG3 and candidate interacting protein Positive control:pGBKT7-53+pGADT7-T. Negative control:pGBKT7-lam+pGADT7. Negative control:pGBKT7-EcBAG3+pGADT7
| [1] |
Qin QW, Chang SF, Ngoh-Lim GH, et al. Characterization of a novel Ranavirus isolated from grouper Epinephelus tauvina[J]. Dis Aquat Org, 2003, 53:1-9.
doi: 10.3354/dao053001 URL |
| [2] |
Liu H, Teng Y, Zheng XC, et al. Complete sequence of a viral nervous necrosis virus(NNV)isolated from red-spotted grouper(Epinephelus akaara)in China[J]. Arch Virol, 2012, 157(4):777-782.
doi: 10.1007/s00705-011-1187-5 URL |
| [3] |
Huang XH, Huang YH, Ouyang ZL, et al. Singapore grouper Iridovirus, a large DNA virus, induces nonapoptotic cell death by a cell type dependent fashion and evokes ERK signaling[J]. Apoptosis, 2011, 16(8):831-845.
doi: 10.1007/s10495-011-0616-y URL |
| [4] |
Li C, Wang LQ, Liu JX, et al. Singapore grouper Iridovirus(SGIV)inhibited autophagy for efficient viral replication[J]. Front Microbiol, 2020, 11:1446.
doi: 10.3389/fmicb.2020.01446 URL |
| [5] |
Huang YH, Zhang Y, Liu ZT, et al. Autophagy participates in lysosomal vacuolation-mediated cell death in RGNNV-infected cells[J]. Front Microbiol, 2020, 11:790.
doi: 10.3389/fmicb.2020.00790 URL |
| [6] |
Li C, Liu JX, Zhang X, et al. Red grouper nervous necrosis virus(RGNNV)induces autophagy to promote viral replication[J]. Fish Shellfish Immunol, 2020, 98:908-916.
doi: 10.1016/j.fsi.2019.11.053 URL |
| [7] |
Huang YH, Huang XH, Yang Y, et al. Involvement of fish signal transducer and activator of transcription 3(STAT3)in nodavirus infection induced cell death[J]. Fish Shellfish Immunol, 2015, 43(1):241-248.
doi: 10.1016/j.fsi.2014.12.031 URL |
| [8] |
Bruno AP, De Simone FI, Iorio V, et al. HIV-1 Tat protein induces glial cell autophagy through enhancement of BAG3 protein levels[J]. Cell Cycle, 2014, 13(23):3640-3644.
doi: 10.4161/15384101.2014.952959 URL |
| [9] |
Rosati A, Graziano V, De Laurenzi V, et al. BAG3:a multifaceted protein that regulates major cell pathways[J]. Cell Death Dis, 2011, 2:e141.
doi: 10.1038/cddis.2011.24 URL |
| [10] |
Shi HY, Xu HD, Li ZJ, et al. BAG3 regulates cell proliferation, migration, and invasion in human colorectal cancer[J]. Tumour Biol, 2016, 37(4):5591-5597.
doi: 10.1007/s13277-015-4403-1 URL |
| [11] |
Behl C. Breaking BAG:the co-chaperone BAG3 in health and disease[J]. Trends Pharmacol Sci, 2016, 37(8):672-688.
doi: 10.1016/j.tips.2016.04.007 URL |
| [12] |
Rosati A, Leone A, Del Valle L, et al. Evidence for BAG3 modulation of HIV-1 gene transcription[J]. J Cell Physiol, 2007, 210(3):676-683.
doi: 10.1002/jcp.20865 URL |
| [13] |
Rosati A, Khalili K, Deshmane SL, et al. BAG3 protein regulates caspase-3 activation in HIV-1-infected human primary microglial cells[J]. J Cell Physiol, 2009, 218(2):264-267.
doi: 10.1002/jcp.21604 URL |
| [14] |
Kyratsous CA, Silverstein SJ. The co-chaperone BAG3 regulates Herpes Simplex Virus replication[J]. PNAS, 2008, 105(52):20912-20917.
doi: 10.1073/pnas.0810656105 pmid: 19088197 |
| [15] | 盛慧, 陈姗姗, 艾聪聪, 等. 利用酵母双杂交技术筛选卵菌效应因子互作蛋白概述[J]. 山东农业大学学报:自然科学版, 2019, 50(3):357-360. |
| Sheng H, Chen SS, Ai CC, et al. A review of interaction proteins of oomycete effectors screened by yeast two hybrid system[J]. J Shandong Agric Univ:Nat Sci Ed, 2019, 50(3):357-360. | |
| [16] |
Brooks D, Naeem F, Stetsiv M, et al. Drosophila NUAK functions with Starvin/BAG3 in autophagic protein turnover[J]. PLoS Genet, 2020, 16(4):e1008700.
doi: 10.1371/journal.pgen.1008700 URL |
| [17] | Meriin AB, Narayanan A, Meng L, et al. Hsp70-Bag3 complex is a hub for proteotoxicity-induced signaling that controls protein aggregation[J]. PNAS, 2018, 115(30):E7043-E7052. |
| [18] | Sherman MY, Gabai V. The role of Bag3 in cell signaling[J]. J Cell Biochem, 2021:jcb. 30111. |
| [19] |
Cirone M. ER stress, UPR activation and the inflammatory response to viral infection[J]. Viruses, 2021, 13(5):798.
doi: 10.3390/v13050798 URL |
| [20] |
Münz C. Autophagy proteins in viral exocytosis and anti-viral immune responses[J]. Viruses, 2017, 9(10):288.
doi: 10.3390/v9100288 URL |
| [21] | Mao JR, Lin E, He L, et al. Autophagy and viral infection[M]// Advances in Experimental Medicine and Biology. Singapore: Springer Singapore, 2019:55-78. |
| [22] |
Iyer K, Chand K, Mitra A, et al. Diversity in heat shock protein families:functional implications in virus infection with a comprehensive insight of their role in the HIV-1 life cycle[J]. Cell Stress Chaperones, 2021, 26(5):743-768.
doi: 10.1007/s12192-021-01223-3 URL |
| [23] |
Rao YL, Wan QY, Su H, et al. ROS-induced HSP70 promotes cytoplasmic translocation of high-mobility group box 1b and stimulates antiviral autophagy in grass carp kidney cells[J]. J Biol Chem, 2018, 293(45):17387-17401.
doi: 10.1074/jbc.RA118.003840 URL |
| [24] |
Klimek C, Jahnke R, Wördehoff J, et al. The Hippo network kinase STK38 contributes to protein homeostasis by inhibiting BAG3-mediated autophagy[J]. Biochim Biophys Acta Mol Cell Res, 2019, 1866(10):1556-1566.
doi: 10.1016/j.bbamcr.2019.07.007 URL |
| [25] |
Sharma A, Batra J, Stuchlik O, et al. Influenza A virus nucleoprotein activates the JNK stress-signaling pathway for viral replication by sequestering host filamin A protein[J]. Front Microbiol, 2020, 11:581867.
doi: 10.3389/fmicb.2020.581867 URL |
| [26] |
Dotson D, Woodruff EA, Villalta F, et al. Filamin A is involved in HIV-1 vpu-mediated evasion of host restriction by modulating tetherin expression[J]. J Biol Chem, 2016, 291(8):4236-4246.
doi: 10.1074/jbc.M115.708123 URL |
| [27] | Liu C, Qin XW, He J, et al. Roles of extracellular matrix components in Tiger frog virus attachment to fathead minnow(Pimephales promelas)cells[J]. Fish Shellfish Immunol, 2020, 107(pt a):9-15. |
| [28] |
Millen S, Gross C, Donhauser N, et al. Collagen IV(COL4A1, COL4A2), a component of the viral biofilm, is induced by the HTLV-1 oncoprotein tax and impacts virus transmission[J]. Front Microbiol, 2019, 10:2439.
doi: 10.3389/fmicb.2019.02439 URL |
| [29] |
Huang SZ, Liu K, Cheng AC, et al. SOCS proteins participate in the regulation of innate immune response caused by viruses[J]. Front Immunol, 2020, 11:558341.
doi: 10.3389/fimmu.2020.558341 URL |
| [30] |
Monie DD, Correia C, Zhang C, et al. Modular network mechanism of CCN1-associated resistance to HSV-1-derived oncolytic immunovirotherapies for glioblastomas[J]. Sci Rep, 2021, 11(1):11198.
doi: 10.1038/s41598-021-90718-1 URL |
| [31] |
Liu Y, Yin W, Wang JW, et al. KRAB-zinc finger protein ZNF268a deficiency attenuates the virus-induced pro-inflammatory response by preventing IKK complex assembly[J]. Cells, 2019, 8(12):1604.
doi: 10.3390/cells8121604 URL |
| [1] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [2] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [3] | LI Shu-peng, YANG Xiu-fen, YUAN Jing-jing, QIU De-wen. Screening of Interacting Proteins with Protein Elicitor Hrip1 and Its Prokaryotic Expression [J]. Biotechnology Bulletin, 2017, 33(6): 182-189. |
| [4] | YUAN Min WANG Li GE Wei-na ZHANG Lan. Studies on the Interaction Between FD and 14-3-3/GRF7 in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2017, 33(5): 117-122. |
| [5] | Huang Zhangke, Zhang Yi’neng, Mo Zhongzhen, Zhou Yuping, Huang Xiaoling, Tian Chang’en. Effects of Mutations in IQ Motif of AtIQM1 on Its Calmodulin Binding [J]. Biotechnology Bulletin, 2014, 0(12): 128-132. |
| [6] | Tang Xiaoli, Wu Wenxian, Han Lei, Yang Xiufen. Screening of Interacting Proteins with Fungal Elicitor PevD1 by Yeast Two Hybrid System and High Expression of Recombinant in E. coli [J]. Biotechnology Bulletin, 2014, 0(10): 113-118. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||