








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 184-194.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1546
Previous Articles Next Articles
LUO Ying( ), TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin(
), TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin( )
)
Received:2021-12-14
Online:2022-10-26
Published:2022-11-11
Contact:
HE Fu-lin
E-mail:yingluo_301@163.com;2339695475@qq.com
LUO Ying, TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin. Cloning and Response Analysis to Abiotic Stress of GbR2R3-MYB1 Gene from Ginkgo biloba[J]. Biotechnology Bulletin, 2022, 38(10): 184-194.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| GbR2R3MYB1-F | AGAAGCTGCTCCTCGTTACAG | 目的基因扩增引物 |
| GbR2R3MYB1-R | GTCCTTGATCAAAATGGGGTA | 目的基因扩增引物 |
| pC007-F | TGTAAAACGACGGCCAGT | pClone007载体引物 |
| pC007-R | CAGGAA ACAGCTATGACC | pClone007载体引物 |
| GFP-F | AGAACACGGGGGACGAGCTCGGTACCATGGGTCGGTCTCCTTGCTG | 亚细胞定位引物 |
| GFP-R | CCCTTGCTCACCATGTCGACTCTAGATACCCGCAGTTGCCTGTAAT | 亚细胞定位引物 |
| Gb18S-F | CGAAGACGATCAGATACCG | qPCR内参基因 |
| Gb18S-R | TCAGCCTTGCGACCATAC | qPCR内参基因 |
| q-R2R3MYB1-F | ACGGCAGCTCCAGCTAATTC | qPCR检测引物 |
| q-R2R3MYB1-R | GTGGTGGCTCTGCTTATGCT | qPCR检测引物 |
Table 1 Primer sequences used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| GbR2R3MYB1-F | AGAAGCTGCTCCTCGTTACAG | 目的基因扩增引物 |
| GbR2R3MYB1-R | GTCCTTGATCAAAATGGGGTA | 目的基因扩增引物 |
| pC007-F | TGTAAAACGACGGCCAGT | pClone007载体引物 |
| pC007-R | CAGGAA ACAGCTATGACC | pClone007载体引物 |
| GFP-F | AGAACACGGGGGACGAGCTCGGTACCATGGGTCGGTCTCCTTGCTG | 亚细胞定位引物 |
| GFP-R | CCCTTGCTCACCATGTCGACTCTAGATACCCGCAGTTGCCTGTAAT | 亚细胞定位引物 |
| Gb18S-F | CGAAGACGATCAGATACCG | qPCR内参基因 |
| Gb18S-R | TCAGCCTTGCGACCATAC | qPCR内参基因 |
| q-R2R3MYB1-F | ACGGCAGCTCCAGCTAATTC | qPCR检测引物 |
| q-R2R3MYB1-R | GTGGTGGCTCTGCTTATGCT | qPCR检测引物 |

Fig.1 Agarose gel electrophoresis of PCR amplified prod-ucts of GbR2R3-MYB1 A:PCR amplification of target fragment of GbR2R3-MYB1. B:Bacteria liquid PCR;M:DL2000 DNA marker;1-3:positive clones of colony PCR identification
Fig. 7 Prediction of secondary structure for GbR2R3-MYB1 protein Blue:Alpha helix. Green:Beta turn. Purple:Random coil. Red:Extended strand. D:Tertiary structure analysis of GbR2R3-MYB1 protein
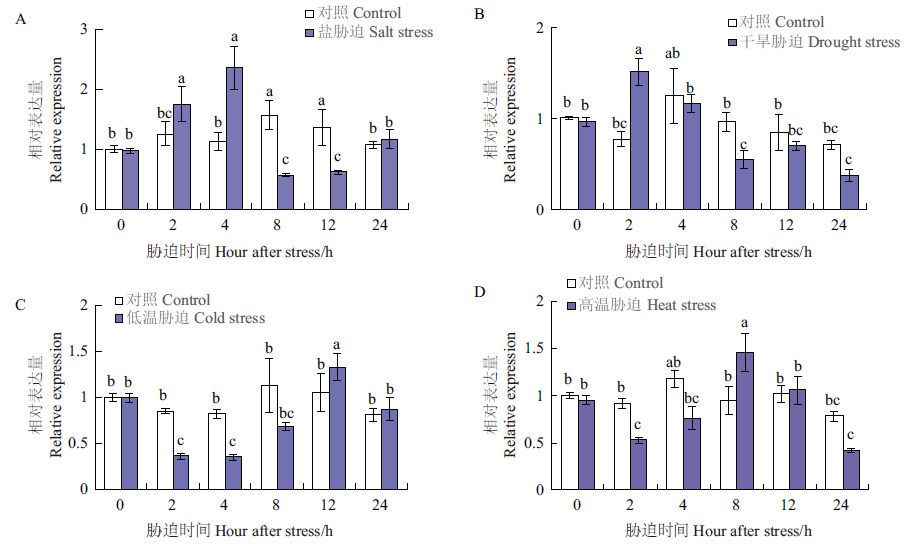
Fig.10 Expression analysis of GbR2R3-MYB1 gene under salt,drought,cold,and heat stresses Each value was calculated as the mean of three samples standard error(SE). The different letters above the bar indicate significant difference(P < 0.05)and very significant difference(P < 0.01)
| [1] | Jin JP, Zhang H, Kong L, et al. PlantTFDB 3. 0:a portal for the functional and evolutionary study of plant transcription factors[J]. Nucleic Acids Res, 2014, 42(Database issue):D1182-D1187. |
| [2] |
Amoutzias GD, Veron AS, Weiner J 3rd, et al. One billion years of bZIP transcription factor evolution:conservation and change in dimerization and DNA-binding site specificity[J]. Mol Biol Evol, 2007, 24(3):827-835.
doi: 10.1093/molbev/msl211 pmid: 17194801 |
| [3] |
Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development[J]. Plant Cell, 2005, 17(3):705-721.
doi: 10.1105/tpc.104.027920 URL |
| [4] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10):573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [5] |
Nesi N, Jond C, Debeaujon I, et al. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed[J]. Plant Cell, 2001, 13(9):2099-2114.
pmid: 11549766 |
| [6] |
Müller D, Schmitz G, Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis[J]. Plant Cell, 2006, 18(3):586-597.
pmid: 16461581 |
| [7] |
Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors:their role in drought response mechanisms[J]. Int J Mol Sci, 2015, 16(7):15811-15851.
doi: 10.3390/ijms160715811 pmid: 26184177 |
| [8] |
Wang Z, Tang J, Hu R, et al. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage(Brassica rapa ssp. pekinensis)reveals their stress and hormone responsive patterns[J]. BMC Genomics, 2015, 16(1):17.
doi: 10.1186/s12864-015-1216-y URL |
| [9] |
Agarwal M, Hao YJ, Kapoor A, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance[J]. J Biol Chem, 2006, 281(49):37636-37645.
doi: 10.1074/jbc.M605895200 pmid: 17015446 |
| [10] |
Xie YP, Chen PX, Yan Y, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple[J]. New Phytol, 2018, 218(1):201-218.
doi: 10.1111/nph.14952 pmid: 29266327 |
| [11] |
Lv Y, Yang M, Hu D, et al. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression[J]. Plant Physiol, 2017, 173(2):1475-1491.
doi: 10.1104/pp.16.01725 pmid: 28062835 |
| [12] |
Liao CC, Zheng Y, Guo Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis[J]. New Phytol, 2017, 216(1):163-177.
doi: 10.1111/nph.14679 URL |
| [13] |
El-Kereamy A, Bi YM, Ranathunge K, et al. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism[J]. PLoS One, 2012, 7(12):e52030.
doi: 10.1371/journal.pone.0052030 URL |
| [14] |
Casaretto JA, El-Kereamy A, Zeng B, et al. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance[J]. BMC Genomics, 2016, 17:312.
doi: 10.1186/s12864-016-2659-5 pmid: 27129581 |
| [15] |
Liu CY, Xie T, Chen CJ, et al. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple(Ananas comosus)[J]. BMC Genomics, 2017, 18(1):503.
doi: 10.1186/s12864-017-3896-y URL |
| [16] | 曹福亮. 中国银杏[M]. 南京: 江苏科学技术出版社, 2002. |
| Cao FL. Chinese Ginkgo[M]. Nanjing: Phoenix Science Press, 2002. | |
| [17] | 陈学森, 张艳敏, 李健, 等. 叶用银杏资源评价及选优的研究[J]. 园艺学报, 1997, 24(3):215-219. |
| Chen XS, Zhang YM, Li J, et al. The evaluation of leaf utilization resources and cultivar selection in Ginkgo biloba[J]. Acta Hortic Sin, 1997, 24(3):215-219. | |
| [18] | 骆鹰, 谢旻, 张超, 等. 水稻Cu/Zn-SOD基因的克隆、表达及生物信息学分析[J]. 分子植物育种, 2018, 16(10):3097-3105. |
| Luo Y, Xie M, Zhang C, et al. Cloning, expression and bioinformatic analysis of Cu/Zn-SOD gene in rice[J]. Mol Plant Breed, 2018, 16(10):3097-3105. | |
| [19] | 谢旻, 骆鹰, 张超, 等. 水稻铜/锌超氧化物歧化酶铜伴侣基因克隆与表达分析[J]. 基因组学与应用生物学, 2017, 36(7):2940-2946. |
| Xie M, Luo Y, Zhang C, et al. Gene cloning and expression analysis of copper chaperone for Cu/Zn superoxide dismutase(OsCCS)in rice[J]. Genom Appl Biol, 2017, 36(7):2940-2946. | |
| [20] | 刘慧春, 马广莹, 朱开元, 等. 牡丹PsDREB转录因子基因的克隆及亚细胞定位[J]. 分子植物育种, 2015, 13(10):2290-2298. |
| Liu HC, Ma GY, Zhu KY, et al. Clonging and subcellular location of a DREB transcription gene of Paeonia suffruticosa[J]. Mol Plant Breed, 2015, 13(10):2290-2298. | |
| [21] | 李泽宏, 袁红慧, 程华, 等. 银杏GbERF1转录因子基因的克隆及亚细胞定位分析[J]. 北方园艺, 2018(3):92-100. |
| Li ZH, Yuan HH, Cheng H, et al. Cloning and subcellular localization analysis of GbERF1 transcription factor in Ginkgo biloba L[J]. North Hortic, 2018(3):92-100. | |
| [22] | 欧阳梦真, 朱磊, 孙治强, 等. 西瓜ClWRKY54基因的克隆、亚细胞定位及表达分析[J]. 中国瓜菜, 2019, 32(12):8-14. |
| Ouyang MZ, Zhu L, Sun ZQ, et al. Cloning, subcellular localization and expression analysis of ClWRKY54 in Citrullus lanatus[J]. China Cucurbits Veg, 2019, 32(12):8-14. | |
| [23] | 杜恬恬, 李会萍, 王博雅, 等. 梁山慈竹DfMYB3基因克隆及启动子分析[J]. 植物研究, 2021, 41(5):729-737. |
| Du TT, Li HP, Wang BY, et al. Cloning and promotor analysis of DfMYB3 from Dendrocalamus farinosus[J]. Bull Bot Res, 2021, 41(5):729-737. | |
| [24] | 彭晶晶, 李凯, 杨靖, 等. 丹参转录因子SmMYB52的基因克隆、表达分析和亚细胞定位[J]. 基因组学与应用生物学, 2021, 40(2):802-808. |
| Peng JJ, Li K, Yang J, et al. Gene clone, expression analysis and subcellular localization of transcription factor SmMYB52 in Salvia miltiorrhiza[J]. Genom Appl Biol, 2021, 40(2):802-808. | |
| [25] | 张顺仓, 冯思念, 顾雯, 等. 丹参中转录因子SmMYB87基因的克隆、亚细胞定位和表达分析[J]. 中草药, 2017, 48(17):3597-3604. |
| Zhang SC, Feng SN, Gu W, et al. Cloning, subcellular localization and expression analysis of a transcription factor gene SmMYB87 in Salvia miltiorrhiza[J]. Chin Tradit Herb Drugs, 2017, 48(17):3597-3604. | |
| [26] |
Li JL, Han GL, Sun CF, et al. Research advances of MYB transcription factors in plant stress resistance and breeding[J]. Plant Signal Behav, 2019, 14(8):1613131.
doi: 10.1080/15592324.2019.1613131 URL |
| [27] |
Zhao YY, Yang ZE, Ding YP, et al. Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis[J]. Plant Sci, 2019, 286:28-36.
doi: 10.1016/j.plantsci.2019.05.021 URL |
| [28] |
Yoo JH, Park CY, Kim JC, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis[J]. J Biol Chem, 2005, 280(5):3697-3706.
doi: 10.1074/jbc.M408237200 URL |
| [29] | Jung C, Seo JS, Han SW, et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis[J]. Plant Physiol, 2008, 146(2):623-635. |
| [30] |
Zhang X, Chen LC, Shi QH, et al. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato[J]. Plant Sci, 2020, 291:110356.
doi: 10.1016/j.plantsci.2019.110356 URL |
| [31] |
Chen N, Yang QL, Pan LJ, et al. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut(Arachis hypogaea L.)[J]. Gene, 2014, 533(1):332-345.
doi: 10.1016/j.gene.2013.08.092 pmid: 24013078 |
| [32] |
Qin YX, Wang MC, Tian YC, et al. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis[J]. Mol Biol Rep, 2012, 39(6):7183-7192.
doi: 10.1007/s11033-012-1550-y URL |
| [33] |
Zhang LC, Zhao GY, Xia C, et al. Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic Arabidopsis[J]. Gene, 2012, 505(1):100-107.
doi: 10.1016/j.gene.2012.05.033 URL |
| [34] |
He YN, Li W, Lv J, et al. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana[J]. J Exp Bot, 2012, 63(3):1511-1522.
doi: 10.1093/jxb/err389 URL |
| [35] | 李春艳, 王曦, 周胜花, 等. 白羊草R2R3-MYB转录因子的挖掘及对干旱胁迫反应的研究[J]. 草地学报, 2020, 28(6):1784-1790. |
| Li CY, Wang X, Zhou SH, et al. Excavation of R2R3-MYB transcription factor and its response to drought stress in Bothriochloa ischaemum[J]. Acta Agrestia Sin, 2020, 28(6):1784-1790. | |
| [36] |
Du YT, Zhao MJ, Wang CT, et al. Identification and characterization of GmMYB118 responses to drought and salt stress[J]. BMC Plant Biol, 2018, 18(1):320.
doi: 10.1186/s12870-018-1551-7 URL |
| [37] |
Xie YP, Chen PX, Yan Y, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple[J]. New Phytol, 2018, 218(1):201-218.
doi: 10.1111/nph.14952 pmid: 29266327 |
| [38] |
An JP, Li R, Qu FJ, et al. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple[J]. Plant J, 2018, 96(3):562-577.
doi: 10.1111/tpj.14050 URL |
| [39] |
Liao Y, Zou HF, Wang HW, et al. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants[J]. Cell Res, 2008, 18(10):1047-1060.
doi: 10.1038/cr.2008.280 URL |
| [40] |
Jacob P, Brisou G, Dalmais M, et al. The seed development factors TT2 and MYB5 regulate heat stress response in Arabidopsis[J]. Genes, 2021, 12(5):746.
doi: 10.3390/genes12050746 URL |
| [41] |
Zhao Y, Tian XJ, Wang F, et al. Characterization of wheat MYB genes responsive to high temperatures[J]. BMC Plant Biol, 2017, 17(1):208.
doi: 10.1186/s12870-017-1158-4 pmid: 29157199 |
| [42] |
Zhang CY, Liu HC, Zhang XS, et al. VcMYB4a, an R2R3-MYB transcription factor from Vaccinium corymbosum, negatively regulates salt, drought, and temperature stress[J]. Gene, 2020, 757:144935.
doi: 10.1016/j.gene.2020.144935 URL |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [3] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [4] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [5] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [6] | LI Yuan-hong, GUO Yu-hao, CAO Yan, ZHU Zhen-zhou, WANG Fei-fei. Research Progress in the Microalgal Growth and Accumulation of Target Products Regulated by Exogenous Phytohormone [J]. Biotechnology Bulletin, 2023, 39(6): 61-72. |
| [7] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [8] | JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum [J]. Biotechnology Bulletin, 2023, 39(5): 217-223. |
| [9] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [10] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [11] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [12] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [13] | YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase [J]. Biotechnology Bulletin, 2023, 39(4): 71-80. |
| [14] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [15] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||