








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 204-212.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0098
Previous Articles Next Articles
WANG Jia-rui1,2( ), SUN Pei-yuan1,2, KE Jin1,2, RAN Bin1,2, LI Hong-you1(
), SUN Pei-yuan1,2, KE Jin1,2, RAN Bin1,2, LI Hong-you1( )
)
Received:2023-02-08
Online:2023-08-26
Published:2023-09-05
Contact:
LI Hong-you
E-mail:wangjiarui803@163.com;lihongyouluod@163.com
WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum[J]. Biotechnology Bulletin, 2023, 39(8): 204-212.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Use |
|---|---|---|
| FtUGT143-F | ATGGACTGGATGGATGAGATGCTA | 基因克隆Gene cloning |
| FtUGT143-R | CTGTCAAATTTCGACACTGTCAAG | 基因克隆Gene cloning |
| qFtUGT143-F | TAAGGAGATGGCTGAAGCTG | 荧光定量Fluorescent quantification |
| qFtUGT143-R | CACCGAACTTGGATATCCGA | 荧光定量Fluorescent quantification |
| qFtUPL7-F | TTCACGGGCACCATTACTGG | 荧光定量内参Fluorescent quantification of internal reference primers |
| qFtUPL7-R | AGGTGGAAGCTGAAGGAAGC | 荧光定量内参Fluorescent quantification of internal reference primers |
Table 1 List of primers'sequences in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Use |
|---|---|---|
| FtUGT143-F | ATGGACTGGATGGATGAGATGCTA | 基因克隆Gene cloning |
| FtUGT143-R | CTGTCAAATTTCGACACTGTCAAG | 基因克隆Gene cloning |
| qFtUGT143-F | TAAGGAGATGGCTGAAGCTG | 荧光定量Fluorescent quantification |
| qFtUGT143-R | CACCGAACTTGGATATCCGA | 荧光定量Fluorescent quantification |
| qFtUPL7-F | TTCACGGGCACCATTACTGG | 荧光定量内参Fluorescent quantification of internal reference primers |
| qFtUPL7-R | AGGTGGAAGCTGAAGGAAGC | 荧光定量内参Fluorescent quantification of internal reference primers |
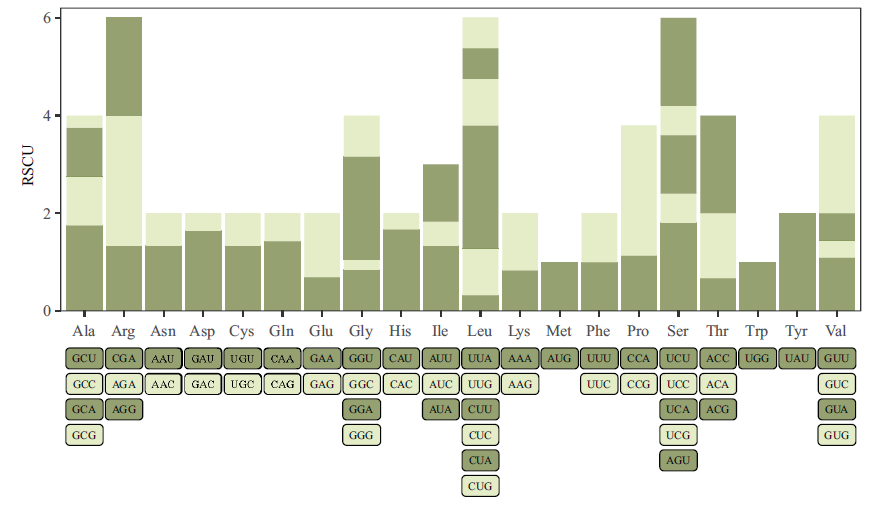
Fig. 2 Analysis of FtUGT143 codon's RSCU values The legend below the abscissa axis corresponds to the RSCU value of the codon above(reflecting the proportional relationship between the actual occurrence times of the codon and the expected occurrence times)
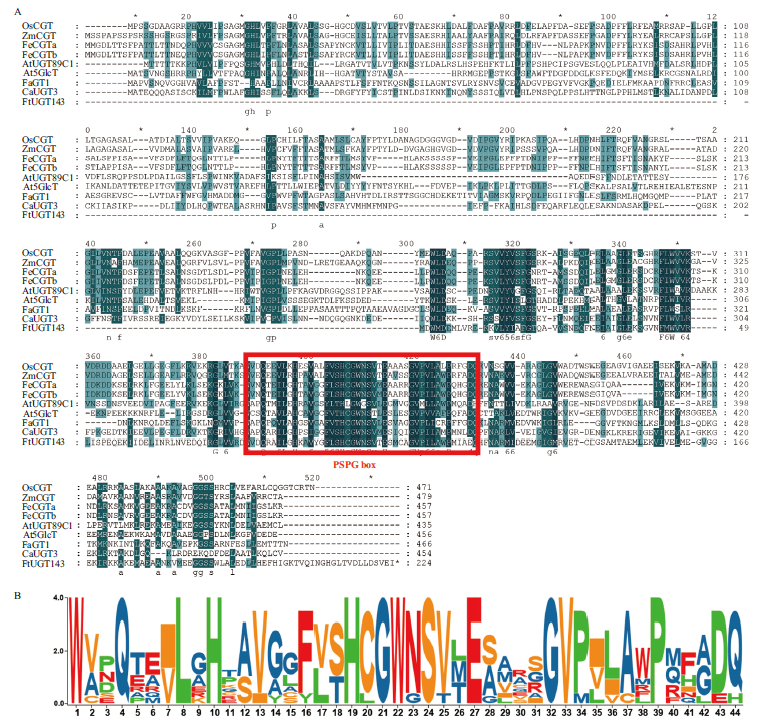
Fig. 3 Sequence multi-alignment of plant UGT protein amino acids and comparative analysis of PSPG box A: Multiple Alignment of amino acid sequences of plant UGT protein(FtUGT143: Fagopyrum tataricum. OsCGT(XP_015641684.1): Oryza sativa; ZmCGT(PWZ07394.1): Zea mays; FeCGTa(A0A0A1HA03.1): Fagopyrum esculentum; FeCGTb(A0A0A1H7N4.1): Fagopyrum esculentum; GmUGT708D1(NP_001347241.1): Glycine max; At5GlcT(NP_193146.1): Arabidopsis thaliana; FaGT1(AAU09442.1): Fragaria × ananassa; CaUGT3(BAH80312.1): Catharanthus roseus). The red box indicates the PSPG box, a conserved sequence motif found in glycosyltransferases. B: Comparative analysis of PSPG box
| 配体小分子 Ligand small molecule | 结合能 Binding energy/(kJ·mol-1) |
|---|---|
| 山奈酚 Kaempferol | - 29.706 4 |
| 表儿茶素 Epicatechin | - 31.796 4 |
| 木犀草素 Luteolin | - 33.890 4 |
| 芹菜素 Apigenin | - 32.216 8 |
Table 2 Docking results of FtUGT143 with four flavonoids metabolizing small molecules
| 配体小分子 Ligand small molecule | 结合能 Binding energy/(kJ·mol-1) |
|---|---|
| 山奈酚 Kaempferol | - 29.706 4 |
| 表儿茶素 Epicatechin | - 31.796 4 |
| 木犀草素 Luteolin | - 33.890 4 |
| 芹菜素 Apigenin | - 32.216 8 |
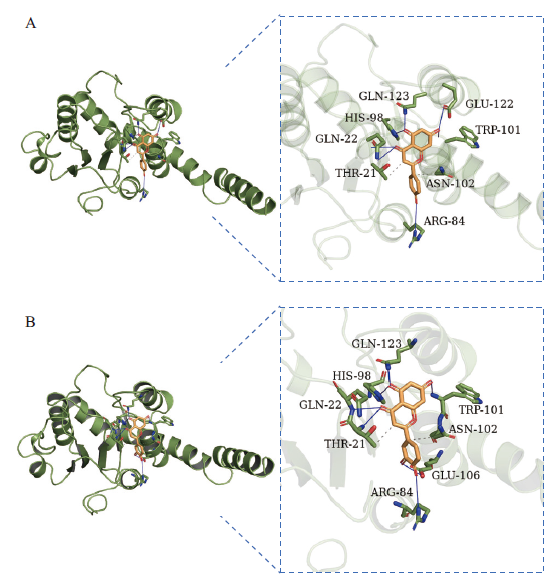
Fig. 5 Schematic diagram of FtUGT143 docking with luteolin and apigenin A: Schematic diagram of FtUGT143 and luteolin docking. B: Schematic diagram of FtUGT143 and apigenin docking
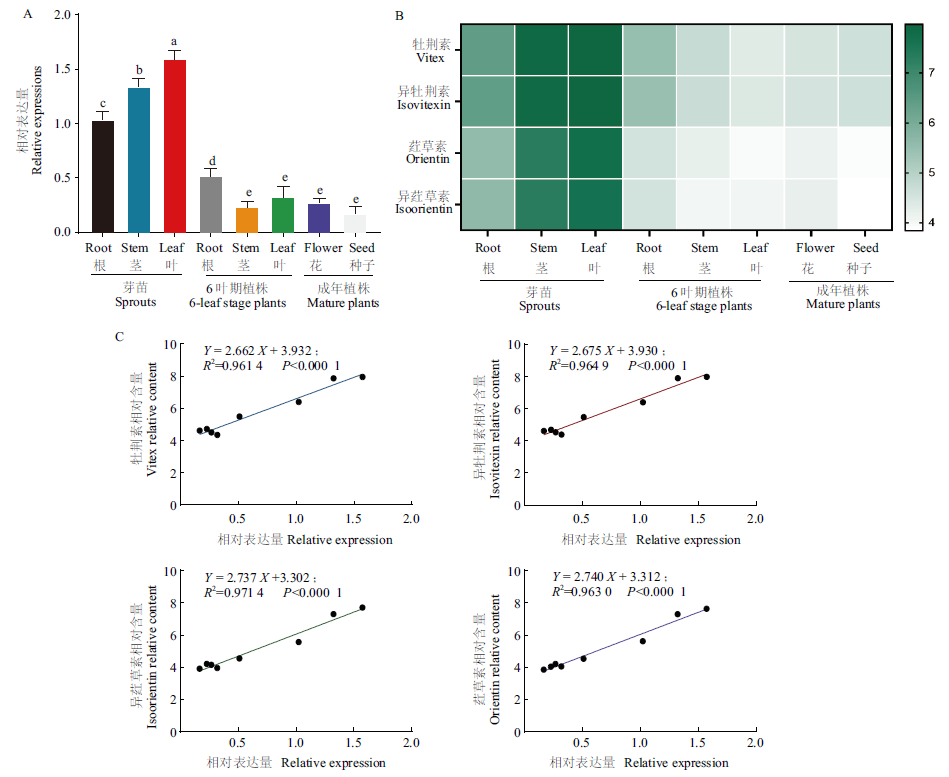
Fig. 6 Correlation between the expressions of FtUGT143 in the different tissue sites of buckwheat and the content of four C-flavonoids A: Expression of FtUGT143 in different tissue sites. B: Relative content of four C-flavonoids in different tissue sites. C: Correlation between the expression of FtUGT143 and the content of four C-flavonoids in different tissue sites
| [1] | Baskar V, Venkatesh R, Ramalingam S. Flavonoids(antioxidants systems)in higher plants and their response to stresses[M]// Antioxidants and Antioxidant Enzymes in Higher Plants. Cham: Springer, 2018: 253-268. |
| [2] | Kumar V, Suman U, et al. Flavonoid secondary metabolite: biosynthesis and role in growth and development in plants[M]// Recent Trends and Techniques in Plant Metabolic Engineering. Singapore: Springer, 2018: 19-45. |
| [3] |
Li HY, Lyu QY, Liu AK, et al. Comparative metabolomics study of Ta-rtary(Fagopyrum tataricum(L.)Gaertn)and common(Fagopyrum esculentum Moench)buckwheat seeds[J]. Food Chem, 2022, 371: 131125.
doi: 10.1016/j.foodchem.2021.131125 URL |
| [4] |
Huda MN, Lu S, Jahan T, et al. Treasure from garden: Bioactive compounds of buckwheat[J]. Food Chem, 2021, 335: 127653.
doi: 10.1016/j.foodchem.2020.127653 URL |
| [5] |
解林峰, 任传宏, 张波, 等. 植物类黄酮生物合成相关UDP-糖基转移酶研究进展[J]. 园艺学报, 2019, 46(9): 1655-1669.
doi: 10.16420/j.issn.0513-353x.2019-0237 |
|
Xie LF, Ren CH, Zhang B, et al. Plant UDP-glycosyltransferases in flavonoids biosynthesis[J]. Acta Hortic Sin, 2019, 46(9): 1655-1669.
doi: 10.16420/j.issn.0513-353x.2019-0237 |
|
| [6] |
Dai LH, Hu YM, Chen CC, et al. Flavonoid C-glycosyltransferases: function, evolutionary relationship, catalytic mechanism and protein engineering[J]. Chembioeng Rev, 2021, 8(1): 15-26.
doi: 10.1002/cben.v8.1 URL |
| [7] | Ren GL, Zhong Y, Ke G, et al. The mechanism of compound Anshen essential oil in the treatment of insomnia was examined by network pharmacology[J]. Evid Based Complement Alternat Med, 2019, 2019: 9241403. |
| [8] | He JB, Zhao P, Hu ZM, et al. Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis[J]. Angewandte Chemie, 2019, 131(33): 11637-11644. |
| [9] |
Wang X, Li CF, Zhou C, et al. Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata[J]. Plant J, 2017, 90(3): 535-546.
doi: 10.1111/tpj.13510 URL |
| [10] |
Mashima K, Hatano M, Suzuki H, et al. Identification and characterization of apigenin 6-C-glucosyltransferase involved in biosynthesis of isosaponarin in wasabi(Eutrema japonicum)[J]. Plant Cell Physiol, 2019, 60(12): 2733-2743.
doi: 10.1093/pcp/pcz164 pmid: 31418788 |
| [11] |
Brazier-Hicks M, Evans KM, Gershater MC, et al. The C-glycosylation of flavonoids in cereals[J]. J Biol Chem, 2009, 284(27): 17926-17934.
doi: 10.1074/jbc.M109.009258 pmid: 19411659 |
| [12] |
Nagatomo Y, Usui S, Ito T, et al. Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M.(buckwheat)cotyledon[J]. Plant J, 2014, 80(3): 437-448.
doi: 10.1111/tpj.2014.80.issue-3 URL |
| [13] |
Yao PF, Deng RY, Huang YJ, et al. Diverse biological effects of glycosyltransferase genes from Tartary buckwheat[J]. BMC Plant Biol, 2019, 19(1): 339.
doi: 10.1186/s12870-019-1955-z pmid: 31382883 |
| [14] |
Yin QG, Han XY, Han ZX, et al. Genome-wide analyses reveals a glucosyltransferase involved in rutin and emodin glucoside biosynthesis in Tartary buckwheat[J]. Food Chem, 2020, 318: 126478.
doi: 10.1016/j.foodchem.2020.126478 URL |
| [15] |
Zhou J, Li CL, Gao F, et al. Characterization of three glucosyltransferase genes in Tartary buckwheat and their expression after cold stress[J]. J Agric Food Chem, 2016, 64(37): 6930-6938.
doi: 10.1021/acs.jafc.6b02064 URL |
| [16] |
Zhang KX, He M, Fan Y, et al. Resequencing of global Tartary buckwheat accessions reveals multiple domestication events and key loci associated with agronomic traits[J]. Genome Biol, 2021, 22(1): 23.
doi: 10.1186/s13059-020-02217-7 pmid: 33430931 |
| [17] |
Xu HT, Jiang ZQ, Lin ZM, et al. FtUGT79A15 is responsible for rutinosylation in flavonoid diglycoside biosynthesis in Fagopyrum tataricum[J]. Plant Physiol Biochem, 2022, 181: 33-41.
doi: 10.1016/j.plaphy.2022.04.004 URL |
| [18] |
Li HY, Lyu QY, Ma C, et al. Metabolite profiling and transcriptome analyses provide insights into the flavonoid biosynthesis in the developing seed of Tartary buckwheat(Fagopyrum tataricum)[J]. J Agric Food Chem, 2019, 67(40): 11262-11276.
doi: 10.1021/acs.jafc.9b03135 URL |
| [19] | 王璐瑗, 荣玉萍, 黄娟, 等. 211份金荞麦收集系根茎黄酮含量的分析评价[J]. 贵州师范大学学报: 自然科学版, 2019, 37(4): 25-30, 48. |
| Wang LY, Rong YP, Huang J, et al. Analysis and evaluation of the flavonoid content of rhizomes of 211 different Golden buckwheat accessions(Fagopyrum cymosum complex)[J]. J Guizhou Norm Univ Nat Sci, 2019, 37(4): 25-30, 48. | |
| [20] |
姚宇, 顾佳珺, 孙超, 等. 植物类黄酮UDP-糖基转移酶研究进展[J]. 生物技术通报, 2022, 38(12): 47-57.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0236 |
|
Yao Y, Gu JJ, Sun C, et al. Advances in plant flavonoids UDP-glycosyltransferase[J]. Biotechnol Bull, 2022, 38(12): 47-57.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0236 |
|
| [21] |
Peng M, Shahzad R, Gul A, et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance[J]. Nat Commun, 2017, 8(1): 1975.
doi: 10.1038/s41467-017-02168-x pmid: 29213047 |
| [22] |
Masada S, Terasaka K, Oguchi Y, et al. Functional and structural characterization of a flavonoid glucoside 1, 6-glucosyltransferase from Catharanthus roseus[J]. Plant Cell Physiol, 2009, 50(8): 1401-1415.
doi: 10.1093/pcp/pcp088 URL |
| [23] | 赵蕾. 毛竹中黄酮-C-糖基转移酶基因的克隆和功能鉴定[D]. 北京: 中国林业科学研究院, 2019. |
| Zhao L. Cloning and functional identification of flavonoid-C-glycosyltransferase genes in Phyllostachys edulis[D]. Beijing: Chinese Academy of Forestry, 2019. | |
| [24] | 付炎, 王于方, 李力更, 等. 天然药物化学史话: 天然产物研究与诺贝尔奖[J]. 中草药, 2016, 47(21): 3749-3765. |
| Fu Y, Wang YF, Li LG, et al. Historical story on natural medicinal chemistry: nature products research and Nobel Prize[J]. Chin Tradit Herb Drugs, 2016, 47(21): 3749-3765. | |
| [25] |
Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family[J]. Trends Plant Sci, 2000, 5(9): 380-386.
doi: 10.1016/s1360-1385(00)01720-9 pmid: 10973093 |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [3] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [4] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [5] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [6] | HOU Rui-ze BAO Yue CHEN Qi-liang MAO Gui-ling WEI Bo-lin HOU Lei-ping LI Mei-lan. Cloning,Expression and Functional Identification of PRR5 Gene in Pakchoi [J]. Biotechnology Bulletin, 2023, 39(10): 128-135. |
| [7] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| [8] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [9] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| [10] | LI Yang, ZHANG Xiao-tian, PIAO Jing-zi, ZHOU Ru-jun, LI Zi-bo, GUAN Hai-wen. Cloning and Bioinformatics Analysis of Blue-light Receptor EaWC 1 Gene in Elsinoë arachidis [J]. Biotechnology Bulletin, 2022, 38(5): 93-99. |
| [11] | HU Qi, HOU Yu-xiang, LI Rui, LI Mei-lan. Cloning and Expression of CYP79B2 Homologous Genes in Brassica rapa ssp. chinensis [J]. Biotechnology Bulletin, 2022, 38(12): 168-174. |
| [12] | GAN Cheng-yan, ZHANG Xin-hui, WANG Sha, FAN Yao-yu-wei, ZHAO Xue-qing, YUAN Zhao-he. Cloning and Functional Study of the PgSPL2 Gene Related to the Development of Pomegranate Flowers [J]. Biotechnology Bulletin, 2022, 38(12): 194-203. |
| [13] | SHENG Xue-qing, ZHAO Nan, LIN Ya-qiu, CHEN Ding-shuang, WANG Rui-long, LI Ao, WANG Yong, LI Yan-yan. Cloning and Expression Analysis of ZNF32 Gene in Goat [J]. Biotechnology Bulletin, 2022, 38(12): 300-311. |
| [14] | FU Wei-jie, KUANG Jie-hua, LUO Jun, HUANG Jian-sheng, CHEN You-ming, CHEN Gang. Gene Cloning of Galectin-8 in Epinephelus fuscoguttatus♀×E. polyphekadion♂ and Its Expression Responses Under Different of Ferulic Acid Level [J]. Biotechnology Bulletin, 2022, 38(12): 312-323. |
| [15] | ZHANG Lin, WEI Zhen-zhen, SONG Cheng-wei, GUO Li-li, GUO Qi, HOU Xiao-gai, WANG Hua-fang. Cloning and Expression Analysis of PoFD Gene from Paeonia ostii ‘Fengdan’ [J]. Biotechnology Bulletin, 2022, 38(11): 104-111. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||