








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 217-223.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1231
Previous Articles Next Articles
JIANG Qing-chun( ), DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu(
), DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu( )
)
Received:2022-10-08
Online:2023-05-26
Published:2023-06-08
Contact:
LIU Zhong-yu
E-mail:2870140725@qq.com;zyliu2004@126.com
JIANG Qing-chun, DU Jie, WANG Jia-cheng, YU Zhi-he, WANG Yun, LIU Zhong-yu. Expression and Function Analysis of Transcription Factor PcMYB2 from Polygonum cuspidatum[J]. Biotechnology Bulletin, 2023, 39(5): 217-223.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ |
|---|---|---|
| 35Sf3 | CGGGATCCCATGGAGTCAAAGATTCA | 55 |
| 35Sr3 PcMYB2-F PcMYB2-R | AAAACTGCAGAGTCCCCCGTGTTCTCTC CGCGGATCCGTATGGGGAGAAGAAA AAAACTGCAGGAAATTTCCGAGCCATCC | 55 |
| qPcActin-F1 | GTCCCTGCCATGTATGTTGC | 58 |
| qPcActin-R1 | ACCATCACCAGAATCCAGCA | |
| qPcMYB2-F | GAGGAAGGGTGGAGAAATGA | 58 |
| qPcMYB2-R | TGCCCTTGTGCTGTATCTGA |
Table 1 Primer sequence information
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ |
|---|---|---|
| 35Sf3 | CGGGATCCCATGGAGTCAAAGATTCA | 55 |
| 35Sr3 PcMYB2-F PcMYB2-R | AAAACTGCAGAGTCCCCCGTGTTCTCTC CGCGGATCCGTATGGGGAGAAGAAA AAAACTGCAGGAAATTTCCGAGCCATCC | 55 |
| qPcActin-F1 | GTCCCTGCCATGTATGTTGC | 58 |
| qPcActin-R1 | ACCATCACCAGAATCCAGCA | |
| qPcMYB2-F | GAGGAAGGGTGGAGAAATGA | 58 |
| qPcMYB2-R | TGCCCTTGTGCTGTATCTGA |
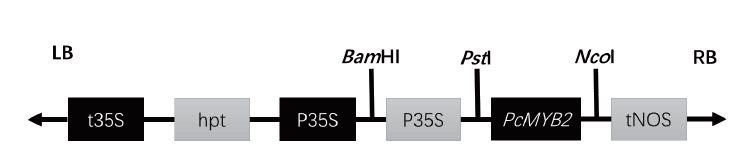
Fig. 2 Structure of pCAMBIA 1380-35S-PcMYB2 vectors LB: Left border of transfer DNA. t35S: Cauliflower mosaic virus CaMV 35S terminator. hpt: Hygromycin resistance gene P35S - Cauliflower mosaic virus CaMV 3SS promoter. tNOS: Nitric oxide synthase gene terminator. RB: Right of transfer DNA boundary
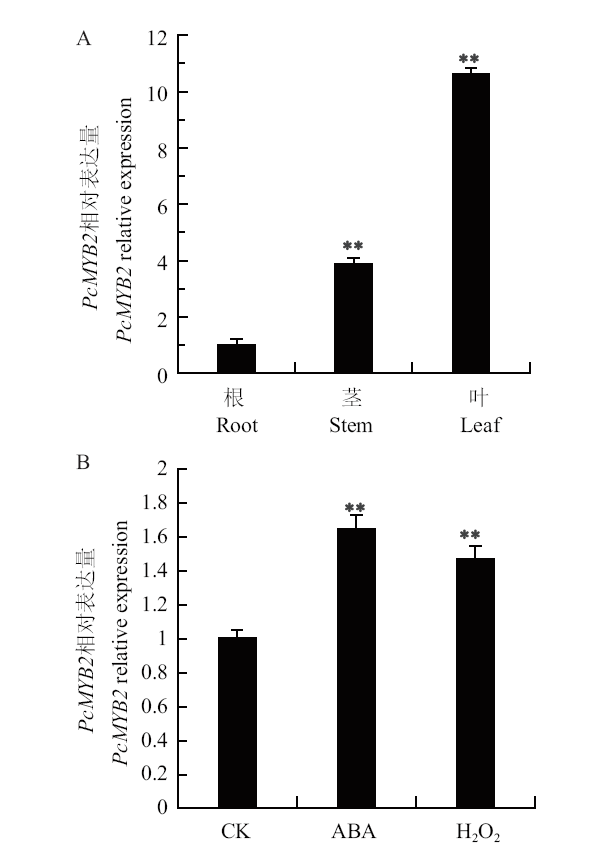
Fig. 5 Expression pattern analysis of PcMYB2 A : Expressions of PcMYB2 gene in different tissues. B : PcMYB2 expressions in leaves under exogenous signaling molecules. ** P < 0.01. The same below

Fig. 6 Verification of recombinant plamid pCAMBIA1380-35S-PcMYB2 M: Marker. 1: Digestion recombinant plasmid by BamH I/Pst I. 2: Digestion recombinant plasmid by BamH I/ Nco I
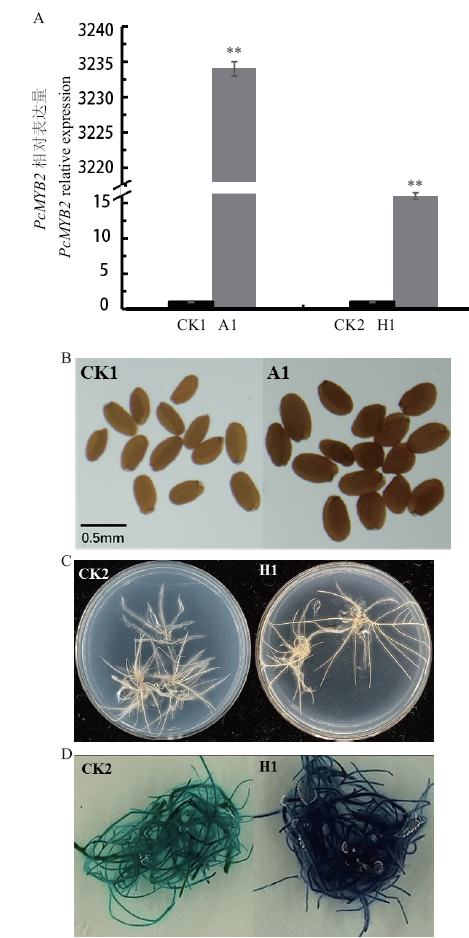
Fig. 7 Identification and phenotypic observation of trans-genic lines A: RT-qPCR identification. B: Observation on the seed coat's color of A. thaliana. C: Hairy roots before DMACA staining. D: Hairy root after DMACA staining. CK1: Wild-type A.thaliana; A1: transgenic A.thaliana; CK2: wild-type hairy roots; H1: transgenic hairy roots
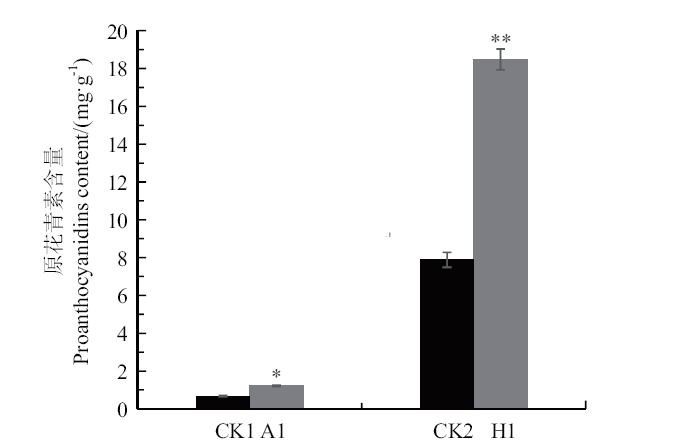
Fig. 8 Determination of proanthocyanidins CK1: Wild-type A.thaliana; A1: transgenic A. thaliana; CK2: wild-type hairy roots; H1: transgenic hairy roots; *P < 0.05
| [1] |
Fan PH, Hostettmann K, Lou HX. Allelochemicals of the invasive neophyte Polygonum cuspidatum Sieb. & Zucc.(Polygonaceae)[J]. Chemoecology, 2010, 20(3): 223-227.
doi: 10.1007/s00049-010-0052-4 URL |
| [2] |
Li X, Liu JL, Chang QX, et al. Antioxidant and antidiabetic activity of proanthocyanidins from Fagopyrum dibotrys[J]. Molecules, 2021, 26(9): 2417.
doi: 10.3390/molecules26092417 URL |
| [3] |
Suganya M, Gnanamangai BM, Ravindran B, et al. Antitumor effect of proanthocyanidin induced apoptosis in human colorectal cancer(HT-29)cells and its molecular docking studies[J]. BMC Chem, 2019, 13(1): 21.
doi: 10.1186/s13065-019-0525-7 pmid: 31384770 |
| [4] |
Wang JY, Zhang BX, Tian ST, et al. Ectopic expression of grape hyacinth R3 MYB repressor MaMYBx affects anthocyanin and proanthocyanidin biosynthesis and epidermal cell differentiation in Arabidopsis[J]. Hortic Environ Biotechnol, 2022, 63(3): 413-423.
doi: 10.1007/s13580-021-00401-7 |
| [5] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [6] |
Zhang JK, Wang YC, Mao ZL, et al. Transcription factor McWRKY71 induced by ozone stress regulates anthocyanin and proanthocyanidin biosynthesis in Malus crabapple[J]. Ecotoxicol Environ Saf, 2022, 232: 113274.
doi: 10.1016/j.ecoenv.2022.113274 URL |
| [7] |
Gil-Muñoz F, Sánchez-Navarro JA, Besada C, et al. MBW complexes impinge on anthocyanidin reductase gene regulation for proanthocyanidin biosynthesis in persimmon fruit[J]. Sci Rep, 2020, 10(1): 3543.
doi: 10.1038/s41598-020-60635-w pmid: 32103143 |
| [8] |
Passeri V, Martens S, Carvalho E, et al. The R2R3MYB VvMYBPA1 from grape reprograms the phenylpropanoid pathway in tobacco flowers[J]. Planta, 2017, 246(2): 185-199.
doi: 10.1007/s00425-017-2667-y pmid: 28299441 |
| [9] | 李晓筱, 徐俊雄, 刘婵, 等. 虎杖MYB转录因子PcMYB2基因的克隆与原核表达[J]. 江苏农业科学, 2018, 46(23): 50-55. |
| Li XX, Xu JX, Liu S, et al. Cloning and prokaryotic expression of MYB transcription factor PcMYB2 gene from Polygonum cuspidatum[J]. Jiangsu Agric Sci, 2018, 46(23): 50-55. | |
| [10] | 房志家, 陈婷, 郝贺龙, 等. 酿酒酵母转化方法的新探索[J]. 实验室研究与探索, 2012, 31(4): 5-8, 78. |
| Fang ZJ, Chen T, Hao HL, et al. Exploration on transformation of saccharomy cescerevisiae[J]. Res Explor Lab, 2012, 31(4): 5-8, 78. | |
| [11] | 张新, 崔广艳, 陈翠娜, 等. 农杆菌介导DR1172基因转化拟南芥[J]. 基因组学与应用生物学, 2012, 31(6): 582-586. |
| Zhang X, Cui GY, Chen CN, et al. Preliminary study on Agrobacterium-mediated transformation of DR1172 gene into Arabidopsis[J]. Genom Appl Biol, 2012, 31(6): 582-586. | |
| [12] | 段梦灵, 李鲁汉, 廖辉, 等. 发根农杆菌介导的虎杖转基因体系优化[J]. 现代农业科技, 2021(4): 46-50. |
| Duan ML, Li LH, Liao H, et al. Optimization of Agrobacterium rhizogenes-mediated Polygonum cuspidatum transgenic system[J]. Mod Agric Sci Technol, 2021(4): 46-50. | |
| [13] |
Verdier J, Zhao J, Torres-Jerez I, et al. MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula[J]. Proc Natl Acad Sci USA, 2012, 109(5): 1766-1771.
doi: 10.1073/pnas.1120916109 pmid: 22307644 |
| [14] |
Wei JB, Yang JF, Jiang WB, et al. Stacking triple genes increased proanthocyanidins level in Arabidopsis thaliana[J]. PLoS One, 2020, 15(6): e0234799.
doi: 10.1371/journal.pone.0234799 URL |
| [15] |
Liu JY, Osbourn A, Ma PD. MYB transcription factors as regulators of phenylpropanoid metabolism in plants[J]. Mol Plant, 2015, 8(5): 689-708.
doi: 10.1016/j.molp.2015.03.012 pmid: 25840349 |
| [16] |
Shan TL, Rong W, Xu HJ, et al. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes[J]. Sci Rep, 2016, 6: 28777.
doi: 10.1038/srep28777 |
| [17] |
Bai QX, Duan BB, Ma JC, et al. Coexpression of PalbHLH1 and PalMYB90 genes from Populus alba enhances pathogen resistance in poplar by increasing the flavonoid content[J]. Front Plant Sci, 2020, 10: 1772.
doi: 10.3389/fpls.2019.01772 URL |
| [18] | Wang S, Wu HL, Cao XX, et al. Tartary buckwheat FtMYB30 transcription factor improves the salt/drought tolerance of transgenic Arabidopsis in an ABA-dependent manner[J]. Physiol Plant, 2022, 174(5): e13781. |
| [19] |
Wei QH, Liu YY, Lan KE, et al. Identification and analysis of MYB gene family for discovering potential regulators responding to abiotic stresses in Curcuma wenyujin[J]. Front Genet, 2022, 13: 894928.
doi: 10.3389/fgene.2022.894928 URL |
| [20] |
Wei QH, Luo QC, Wang RB, et al. A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants[J]. Front Plant Sci, 2017, 8: 1374.
doi: 10.3389/fpls.2017.01374 pmid: 28848578 |
| [21] | 王霜, 雒晓鹏, 姚英俊, 等. 苦荞R2R3-MYB转录因子调控原花青素生物合成的研究[J]. 西北植物学报, 2019, 39(11): 1911-1918. |
| Wang S, Luo XP, Yao YJ, et al. Characterization of an R2R3-MYB transcription factor involved in the synthesis of proanthocyanidins from Tartary buckwheat[J]. Acta Bot Boreali Occidentalia Sin, 2019, 39(11): 1911-1918. | |
| [22] |
Liu CG, Jun JH, Dixon RA. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula[J]. Plant Physiol, 2014, 165(4): 1424-1439.
doi: 10.1104/pp.114.241877 URL |
| [23] |
Jin Z, Jiang WB, Luo YJ, et al. Analyses on flavonoids and transcriptome reveals key MYB gene for proanthocyanidins regulation in Onobrychis viciifolia[J]. Front Plant Sci, 2022, 13: 941918.
doi: 10.3389/fpls.2022.941918 URL |
| [24] |
An JP, Xu RR, Liu X, et al. Jasmonate induces biosynthesis of anthocyanin and proanthocyanidin in apple by mediating the JAZ1-TRB1-MYB9 complex[J]. Plant J, 2021, 106(5): 1414-1430.
doi: 10.1111/tpj.v106.5 URL |
| [25] |
张慧文, 张玉, 马超美. 原花青素的研究进展[J]. 食品科学, 2015, 36(5): 296-304.
doi: 10.7506/spkx1002-6630-201505052 |
| Zhang HW, Zhang Y, Ma CM. Progress in procyanidins research[J]. Food Sci, 2015, 36(5): 296-304. | |
| [26] |
Luo XP, Zhao HX, Yao PF, et al. An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and proanthocyanidins from Tartary buckwheat[J]. J Plant Growth Regul, 2018, 37(1): 76-84.
doi: 10.1007/s00344-017-9709-3 URL |
| [27] |
Akagi T, Ikegami A, Yonemori K. DkMyb2 wound-induced transcription factor of persimmon(Diospyros kaki Thunb.), contributes to proanthocyanidin regulation[J]. Planta, 2010, 232(5): 1045-1059.
doi: 10.1007/s00425-010-1241-7 URL |
| [1] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [2] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [3] | YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum [J]. Biotechnology Bulletin, 2023, 39(5): 92-102. |
| [4] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [5] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [6] | WANG Tao, QI Si-yu, WEI Chao-ling, WANG Yi-qing, DAI Hao-min, ZHOU Zhe, CAO Shi-xian, ZENG Wen, SUN Wei-jiang. Expression Analysis and Interaction Protein Validation of CsPPR and CsCPN60-like in Albino Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(3): 218-231. |
| [7] | PANG Qiang-qiang, SUN Xiao-dong, ZHOU Man, CAI Xing-lai, ZHANG Wen, WANG Ya-qiang. Cloning of BrHsfA3 in Chinese Flowering Cabbage and Its Responses to Heat Stress [J]. Biotechnology Bulletin, 2023, 39(2): 107-115. |
| [8] | MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(2): 96-106. |
| [9] | GE Wen-dong, WANG Teng-hui, MA Tian-yi, FAN Zhen-yu, WANG Yu-shu. Genome-wide Identification of the PRX Gene Family in Cabbage(Brassica oleracea L. var. capitata)and Expression Analysis Under Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 252-260. |
| [10] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [11] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [12] | YOU Chui-huai, XIE Jin-jin, ZHANG Ting, CUI Tian-zhen, SUN Xin-lu, ZANG Shou-jian, WU Yi-ning, SUN Meng-yao, QUE You-xiong, SU Ya-chun. Identification of the Lipoxygenase Gene GeLOX1 and Expression Analysis Under Low Temperature Stress in Gelsmium elegans [J]. Biotechnology Bulletin, 2023, 39(11): 318-327. |
| [13] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [14] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| [15] | CHEN Ying, WANG Yi-lei, ZOU Peng-fei. Cloning and Expression Analysis of TRAF6 from Large Yellow Croaker Larimichthys crocea [J]. Biotechnology Bulletin, 2022, 38(8): 233-243. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||