








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 205-216.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0812
Previous Articles Next Articles
LIU Kui( ), LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo(
), LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo( ), ZHANG Ling-yun(
), ZHANG Ling-yun( )
)
Received:2022-07-02
Online:2023-05-26
Published:2023-06-08
Contact:
CAO Yi-bo, ZHANG Ling-yun
E-mail:742174371@qq.com;caoyibo@bjfu.edu.cn;lyzhang@bjfu.edu.cn
LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii[J]. Biotechnology Bulletin, 2023, 39(5): 205-216.
| 用途Use | 引物名称Primer name | 序列Sequence |
|---|---|---|
| RT-qPCR | AtActin-F | GGTAACATTGTGCTCAGTGGTGG |
| AtActin-R | AACGACCTTAATCTTCATGCTGC | |
| PwEF1α-RT-F | AACTGGAGAAGGAACCCAAG | |
| PwEF1α-RT-R | AACGACCCAATGGAGGATAC | |
| PwMBF1c-RT-F | ATGCCGAGCCGAACGAA | |
| PwMBF1c-RT-R | AACCGCCTTGGGATCACG | |
| 载体构建 Vector construction | pEASY-T1-PwMBF1c-F | GAAGGATGCCGAGCCGAA |
| pEASY-T1-PwMBF1c-R | GATGACGACAAGCAATTAGTGAGA | |
| pGBKT7-PwMBF1c-F | CCGGAATTCATGCCGAGCCGAACGAAC | |
| pGBKT7-PwMBF1c-R | TCCCCCGGGGGTGGACTGATGACGACAAGCAATTA | |
| pGBKT7-PwMBF1c-C-F | CCGGAATTCGATTCGACATGCTATACAGAAGGC | |
| pGBKT7-PwMBF1c-C-R | TCCCCCGGGGGTGGACTGATGACGACAAGCAATTA | |
| pGBKT7-PwMBF1c-N-F | CCGGAATTCATGCCGAGCCGAACGAAC | |
| pGBKT7-PwMBF1c-N-R | TCCCCCGGGGGGCCTTCTGTATAGCATGTCGAATC |
Table 1 Primer sequences
| 用途Use | 引物名称Primer name | 序列Sequence |
|---|---|---|
| RT-qPCR | AtActin-F | GGTAACATTGTGCTCAGTGGTGG |
| AtActin-R | AACGACCTTAATCTTCATGCTGC | |
| PwEF1α-RT-F | AACTGGAGAAGGAACCCAAG | |
| PwEF1α-RT-R | AACGACCCAATGGAGGATAC | |
| PwMBF1c-RT-F | ATGCCGAGCCGAACGAA | |
| PwMBF1c-RT-R | AACCGCCTTGGGATCACG | |
| 载体构建 Vector construction | pEASY-T1-PwMBF1c-F | GAAGGATGCCGAGCCGAA |
| pEASY-T1-PwMBF1c-R | GATGACGACAAGCAATTAGTGAGA | |
| pGBKT7-PwMBF1c-F | CCGGAATTCATGCCGAGCCGAACGAAC | |
| pGBKT7-PwMBF1c-R | TCCCCCGGGGGTGGACTGATGACGACAAGCAATTA | |
| pGBKT7-PwMBF1c-C-F | CCGGAATTCGATTCGACATGCTATACAGAAGGC | |
| pGBKT7-PwMBF1c-C-R | TCCCCCGGGGGTGGACTGATGACGACAAGCAATTA | |
| pGBKT7-PwMBF1c-N-F | CCGGAATTCATGCCGAGCCGAACGAAC | |
| pGBKT7-PwMBF1c-N-R | TCCCCCGGGGGGCCTTCTGTATAGCATGTCGAATC |

Fig. 3 Subcellular localization of PwMBF1c GFP: pCM1205 empty vector. PwMBF1c-GFP: Fusion protein of PwMBF1c and GFP tag. A: Subcellular localization of PwMBF1c in tobacco leaves. B: Subcellular localization of PwMBF1c in onion. The bar in the image: 50 µm
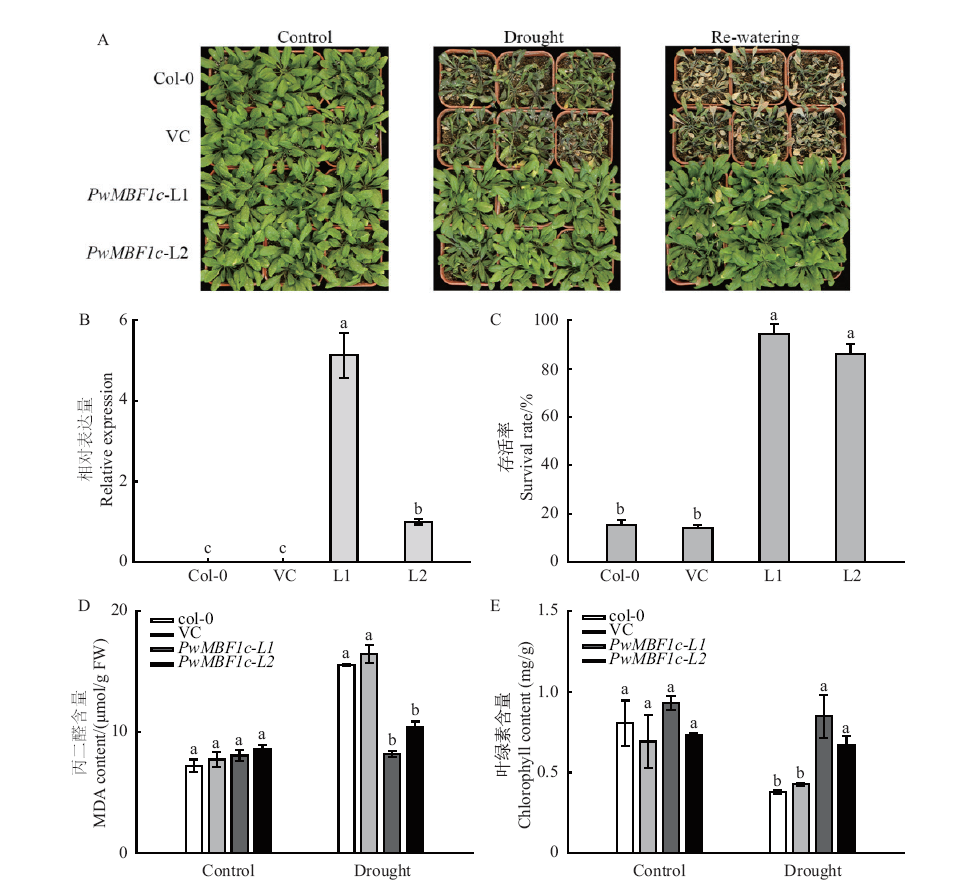
Fig. 5 Overexpression of PwMBF1c significantly improved the drought tolerance of Arabidopsis Col-0: Wild type; VC: empty vector; L1, L2: overexpressing lines of PwMBF1c. A: Phenotypes of different PwMBF1c transgenic lines under drought treatment. B: Relative expressions of PwMBF1c in different lines. C: Statistics of survival rates after drought treatment. D, E: MDA content and chlorophyll content of isolated leaves under drought treatment

Fig. 6 Germination rates of different lines of Arabidopsis under drought stress A: Germination status of different lines treated with 0/100/200/300 mmol/L mannitol. B-E: Germination rates of different lines treated with 0,100,200, and 300 mmol/L mannitol.

Fig. 7 Root length of overexpressing PwMBF1c in Arabidopsis A: Root length growths of different lines treated with mannitol. B: Root length statistics of different lines treated with mannitol
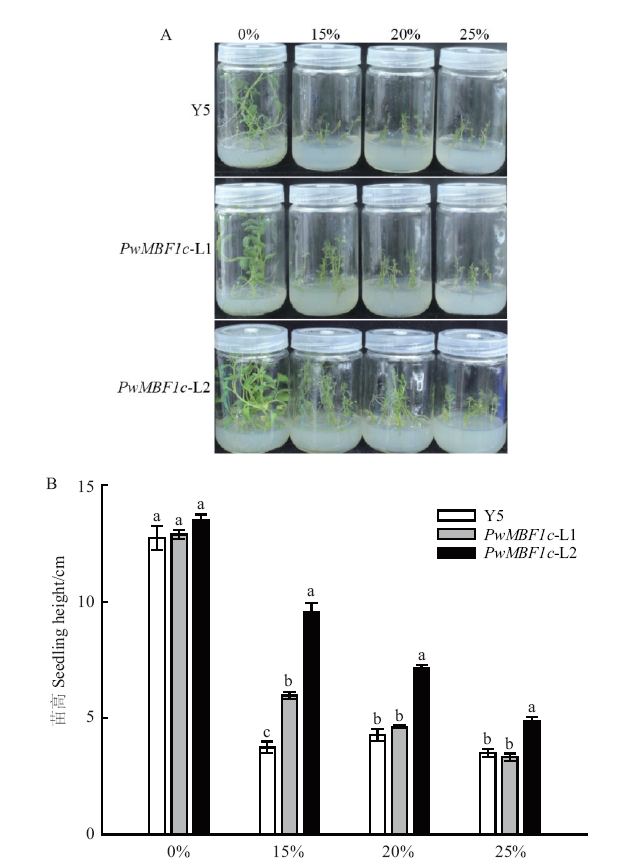
Fig. 8 Drought phenotypes of different potato lines Y5: Wild type. A: Growth status of different lines under PEG treatment. B: Seedling heights of different lines under PEG treatments
| [1] |
Hommel M, Khalil-Ahmad Q, Jaimes-Miranda F, et al. Over-expression of a chimeric gene of the transcriptional co-activator MBF1 fused to the EAR repressor motif causes developmental alteration in Arabidopsis and tomato[J]. Plant Sci, 2008, 175(1-2): 168-177.
doi: 10.1016/j.plantsci.2008.01.019 URL |
| [2] |
Ray S, Dansana PK, Giri J, et al. Modulation of transcription factor and metabolic pathway genes in response to water-deficit stress in rice[J]. Funct Integr Genomics, 2011, 11(1): 157-178.
doi: 10.1007/s10142-010-0187-y URL |
| [3] |
Guo WL, Chen RG, Du XH, et al. Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1[J]. BMC Plant Biol, 2014, 14: 138.
doi: 10.1186/1471-2229-14-138 |
| [4] |
Zhao Q, He L, Wang B, et al. Overexpression of a multiprotein bridging factor 1 gene DgMBF1 improves the salinity tolerance of Chrysanthemum[J]. Int J Mol Sci, 2019, 20(10): 2453.
doi: 10.3390/ijms20102453 URL |
| [5] |
Suzuki N, Rizhsky L, Liang HJ, et al. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c[J]. Plant Physiol, 2005, 139(3): 1313-1322.
doi: 10.1104/pp.105.070110 pmid: 16244138 |
| [6] |
Wang XL, Du Y, Yu DQ. Trehalose phosphate synthase 5-dependent trehalose metabolism modulates basal defense responses in Arabidopsis thaliana[J]. J Integr Plant Biol, 2019, 61(4): 509-527.
doi: 10.1111/jipb.v61.4 URL |
| [7] |
Qin DD, Wang F, Geng XL, et al. Overexpression of heat stress-responsive TaMBF1c, a wheat(Triticum aestivum L.) Multiprotein Bridging Factor, confers heat tolerance in both yeast and rice[J]. Plant Mol Biol, 2015, 87(1-2): 31-45.
doi: 10.1007/s11103-014-0259-9 URL |
| [8] |
Tian XJ, Qin Z, Zhao Y, et al. Stress Granule-associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat(Triticum aestivum)[J]. New Phytol, 2022, 233(4): 1719-1731.
doi: 10.1111/nph.v233.4 URL |
| [9] |
Saleh O, Harb J, Karrity A, et al. Identification of differentially expressed genes in two grape varieties cultivated in semi-arid and temperate regions from West-Bank, Palestine[J]. Agri Gene, 2018, 7: 34-42.
doi: 10.1016/j.aggene.2017.11.001 URL |
| [10] |
Xu Y, Huang BR. Transcriptomic analysis reveals unique molecular factors for lipid hydrolysis, secondary cell-walls and oxidative protection associated with thermotolerance in perennial grass[J]. BMC Genomics, 2018, 19(1): 70.
doi: 10.1186/s12864-018-4437-z pmid: 29357827 |
| [11] |
李小冬, 尚以顺, 武语迪, 等. 紫花苜蓿MsMBF1c基因在拟南芥中表达提高转基因植株的耐热性[J]. 草业学报, 2019, 28(10): 187-198.
doi: 10.11686/cyxb2018739 |
| Li XD, Shang YS, Wu YD, et al. Overexpression of Medicago sativa Multi protein Bridging Factor 1c(MsMBF1c)enhances thermotolerance of Arabidopsis[J]. Acta Prataculturae Sin, 2019, 28(10): 187-198. | |
| [12] |
Yan Q, Hou HM, Singer SD, et al. The grape VvMBF1 gene improves drought stress tolerance in transgenic Arabidopsis thaliana[J]. Plant Cell Tissue Organ Cult, 2014, 118(3): 571-582.
doi: 10.1007/s11240-014-0508-2 URL |
| [13] | 齐瑞, 郭星, 赵阳, 等. 白龙江珍稀濒危植物大果青杄群落物种多样性特征分析[J]. 西北林学院学报, 2017, 32(2): 161-164. |
| Qi R, Guo X, Zhao Y, et al. Species diversity characteristics of the community of rare and endangered Picea neoveitchii in the Bailongjiang River[J]. J Northwest For Univ, 2017, 32(2): 161-164. | |
| [14] |
Zhang H, Shi Y, Liu XR, et al. Transgenic creeping bentgrass plants expressing a Picea wilsonii dehydrin gene(PicW)demonstrate improved freezing tolerance[J]. Mol Biol Rep, 2018, 45(6): 1627-1635.
doi: 10.1007/s11033-018-4304-7 pmid: 30105551 |
| [15] |
Li LL, Yu YL, Wei J, et al. Homologous HAP5 subunit from Picea wilsonii improved tolerance to salt and decreased sensitivity to ABA in transformed Arabidopsis[J]. Planta, 2013, 238(2): 345-356.
doi: 10.1007/s00425-013-1894-0 URL |
| [16] | 苗雅慧, 鞠丹, 梁珂豪, 等. 青杄转录因子基因PwNF-YB8的克隆与功能分析[J]. 林业科学, 2021, 57(5): 77-92. |
| Miao YH, Ju D, Liang KH, et al. Cloning and functional analysis of transcription factor gene PwNF-YB8 from Picea wilsonii[J]. Sci Silvae Sin, 2021, 57(5): 77-92. | |
| [17] | 刘峻玲, 梁珂豪, 苗雅慧, 等. 青杄PwUSP1基因特征及对干旱和盐胁迫的响应[J]. 北京林业大学学报, 2020, 42(10): 62-70. |
| Liu JL, Liang KH, Miao YH, et al. Characteristics of PwUSP1 in Picea wilsonii and its response to drought and salt stress[J]. J Beijing For Univ, 2020, 42(10): 62-70. | |
| [18] | 崔潇月. 青杄盐胁迫响应基因筛选及功能验证体系的建立[D]. 北京: 北京林业大学, 2018. |
| Cui XY. Screening of salt stress response genes in Picea wilsonii and establishment of gene function verification system[D]. Beijing: Beijing Forestry University, 2018. | |
| [19] |
刘亚静, 马齐军, 李浩浩, 等. 苹果MdCBL3基因的克隆与功能鉴定[J]. 园艺学报, 2017, 44(1): 1-10.
doi: 10.16420/j.issn.0513-353x.2016-0489 |
| Liu YJ, Ma QJ, Li HH, et al. Molecular cloning and functional characterization of MdCBL3 in apple[J]. Acta Hortic Sin, 2017, 44(1): 1-10. | |
| [20] |
Zhang FJ, Xie YH, Jiang H, et al. The ankyrin repeat-containing protein MdANK2B regulates salt tolerance and ABA sensitivity in Malus domestica[J]. Plant Cell Rep, 2021, 40(2): 405-419.
doi: 10.1007/s00299-020-02642-9 |
| [21] |
Li FQ, Ueda H, et al. Multiprotein bridging factor 1(MBF1)is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein[J]. Proc Natl Acad Sci USA, 1997, 94(14): 7251-7256.
doi: 10.1073/pnas.94.14.7251 URL |
| [22] |
Brendel C, Gelman L, Auwerx J. Multiprotein bridging factor-1(MBF-1)is a cofactor for nuclear receptors that regulate lipid metabolism[J]. Mol Endocrinol, 2002, 16(6): 1367-1377.
pmid: 12040021 |
| [23] |
Wang YY, Wei XL, Huang J, et al. Modification and functional adaptation of the MBF1 gene family in the lichenized fungus Endocarpon pusillum under environmental stress[J]. Sci Rep, 2017, 7(1): 16333.
doi: 10.1038/s41598-017-16716-4 |
| [24] |
Alavilli H, Lee H, Park M, et al. Antarctic moss multiprotein bridging factor 1c overexpression in Arabidopsis resulted in enhanced tolerance to salt stress[J]. Front Plant Sci, 2017, 8: 1206.
doi: 10.3389/fpls.2017.01206 pmid: 28744295 |
| [25] |
Uji T, Sato R, Mizuta H, et al. Changes in membrane fluidity and phospholipase D activity are required for heat activation of PyMBF1 in Pyropia yezoensis(Rhodophyta)[J]. J Appl Phycol, 2013, 25(6): 1887-1893.
doi: 10.1007/s10811-013-0006-7 URL |
| [26] |
Zhang Y, Zhang G, Dong YL, et al. Cloning and characterization of a MBF1 transcriptional coactivator factor in wheat induced by stripe rust pathogen[J]. Acta Agronomica Sinica, 2009, 35(1): 11-17.
doi: 10.3724/SP.J.1006.2009.00011 URL |
| [27] |
Sugikawa Y, Ebihara S, Tsuda K, et al. Transcriptional coactivator MBF1s from Arabidopsis predominantly localize in nucleolus[J]. J Plant Res, 2005, 118(6): 431-437.
pmid: 16283071 |
| [28] |
Kim GD, Cho YH, Yoo SD. Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature[J]. Biochem Biophys Res Commun, 2015, 457(2): 213-220.
doi: 10.1016/j.bbrc.2014.12.090 URL |
| [29] | 赵楠. 菊花CmMBF1c基因调控耐涝性的分子机制研究[D]. 南京: 南京农业大学, 2019. |
| Zhao N. Molecular mechanisms of CmMBF1c involved in chrysanthemum waterlogging tolerance[D]. Nanjing: Nanjing Agricultural University, 2019. | |
| [30] |
Hao YJ, Wei W, Song QX, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants[J]. Plant J, 2011, 68(2): 302-313.
doi: 10.1111/j.1365-313X.2011.04687.x URL |
| [31] |
Rizhsky L, Liang HJ, Shuman J, et al. When defense pathways collide. the response of Arabidopsis to a combination of drought and heat stress[J]. Plant Physiol, 2004, 134(4): 1683-1696.
doi: 10.1104/pp.103.033431 pmid: 15047901 |
| [32] |
Sah SK, Reddy KR, Li JX. Abscisic acid and abiotic stress tolerance in crop plants[J]. Front Plant Sci, 2016, 7: 571.
doi: 10.3389/fpls.2016.00571 pmid: 27200044 |
| [33] |
Li YM, Sun XR, You JL, et al. Transcriptional analysis of the early ripening of ‘Summer Black’ grape in response to abscisic acid treatment[J]. Sci Hortic, 2022, 299: 111054.
doi: 10.1016/j.scienta.2022.111054 URL |
| [34] |
Baloglu MC, Inal B, Kavas M, et al. Diverse expression pattern of wheat transcription factors against abiotic stresses in wheat species[J]. Gene, 2014, 550(1): 117-122.
doi: 10.1016/j.gene.2014.08.025 pmid: 25130909 |
| [35] |
Katano K, Kataoka R, Fujii M, et al. Differences between seedlings and flowers in anti-ROS based heat responses of Arabidopsis plants deficient in cyclic nucleotide gated channel 2[J]. Plant Physiol Biochem, 2018, 123: 288-296.
doi: 10.1016/j.plaphy.2017.12.021 URL |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [3] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [4] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [5] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [6] | WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation [J]. Biotechnology Bulletin, 2023, 39(4): 59-70. |
| [7] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [8] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [9] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [10] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [11] | YAN Xiong-ying, WANG Zhen, WANG Xia, YANG Shi-hui. Microbial Sulfur Metabolism and Stress Resistance [J]. Biotechnology Bulletin, 2023, 39(11): 150-167. |
| [12] | ZHANG Hong-hong, FANG Xiao-feng. Advances in the Regulation of Stress Sensing and Responses by Phase Separation in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 44-53. |
| [13] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [14] | LIU Yuan-yuan, WEI Chuan-zheng, XIE Yong-bo, TONG Zong-jun, HAN Xing, GAN Bing-cheng, XIE Bao-gui, YAN Jun-jie. Characteristics of Class II Peroxidase Gene Expression During Fruiting Body Development and Stress Response in Flammulina filiformis [J]. Biotechnology Bulletin, 2023, 39(11): 340-349. |
| [15] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||