








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 49-57.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0106
Previous Articles Next Articles
LIU Jia-hui1,2( ), LIU Ye2, HUA Er-bing1(
), LIU Ye2, HUA Er-bing1( ), WANG Meng2(
), WANG Meng2( )
)
Received:2023-02-13
Online:2023-09-26
Published:2023-10-24
Contact:
HUA Er-bing, WANG Meng
E-mail:liujiah@tib.cas.cn;huarb@tust.edu.cn;wangmeng@tib.cas.cn
LIU Jia-hui, LIU Ye, HUA Er-bing, WANG Meng. PAM Extension of Cytosine Base Editing Tool in Corynebacterium glutamicum[J]. Biotechnology Bulletin, 2023, 39(9): 49-57.
| Name | Source | |
|---|---|---|
| Plasmid | pXMJ19TS | Lab stock |
| pnCas9(D10A)-AIDTS | [15] | |
| pgRNA-ccdB | [15] | |
| pnCas9-Sc++(D10A)-AID TS | This study | |
| pnCas9-SpG(D10A)-AID TS | This study | |
| pnCas9-SpRY(D10A)-AID TS | This study | |
| Strain | Escherichia coli DH5α | Lab stock |
| C. glutamicum ATCC 13032 | Lab stock | |
Table 1 Plasmids and strains used in this study
| Name | Source | |
|---|---|---|
| Plasmid | pXMJ19TS | Lab stock |
| pnCas9(D10A)-AIDTS | [15] | |
| pgRNA-ccdB | [15] | |
| pnCas9-Sc++(D10A)-AID TS | This study | |
| pnCas9-SpG(D10A)-AID TS | This study | |
| pnCas9-SpRY(D10A)-AID TS | This study | |
| Strain | Escherichia coli DH5α | Lab stock |
| C. glutamicum ATCC 13032 | Lab stock | |
| Primer name | Sequence(5'-3') |
|---|---|
| ScD10A-1F | TAAGCTTAAAGGAGTTGAGAATGGAGAAGAAATACTCTATCGGCC |
| ScD10A-1R | GGAAGTCCAGGATGGTCTTGCCGGACTGCT |
| ScD10A-2F | CAAGACCATCCTGGACTTCCTGAAGTCCGATG |
| ScD10A-2R | ACCTTGCGTTTCTTCTTTGGATCGCCGCCCAACTGAGAGA |
| ScD10A-3F | CCAAAGAAGAAACGCAAGGTCG |
| S-3R | CCGCCGCCAAGTGATTCTTAG |
| ScD10A-4F | CTAAGAATCACTTGGCGGCGG |
| ScD10A-4R | TCTCAACTCCTTTAAGCTTAATTAA |
| SpG-1F | TAAGCTTAAAGGAGTTGAGAATGGAT |
| SpG-1R | AAAACCACCATATTTTTTTGGATCC |
| SpG-2F | CAAAAAAATATGGTGGTTTTCTGTGGCCAACGGTAGCTTAT |
| SpG-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTACGATCAATTGTT |
| S-3F | TCTACAAAAGAAGTTTTAGATGCCAC |
| SpG-4F | CTAAGAATCACTTGGCGGC |
| SpG-4R | TCTCAACTCCTTTAAGCTTAATTAATTCT |
| SpRY-1F | AGCGGAACGCACTCGTCTCA |
| SpRY-1R | AATTGACTCCTTGGAGAATCCGC |
| SpRY-2F | GATTCTCCAAGGAGTCAATTCGCCCAAAAAGAAATTCGGACA |
| SpRY-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTTGGATCAATTGTT |
| SpRY-4F | CTAAGAATCACTTGGCGGCGGGTG |
| SpRY-4R | TGAGACGAGTGCGTTCCGCTGTCTC |
Table 2 Primers for plasmids construction used in this study
| Primer name | Sequence(5'-3') |
|---|---|
| ScD10A-1F | TAAGCTTAAAGGAGTTGAGAATGGAGAAGAAATACTCTATCGGCC |
| ScD10A-1R | GGAAGTCCAGGATGGTCTTGCCGGACTGCT |
| ScD10A-2F | CAAGACCATCCTGGACTTCCTGAAGTCCGATG |
| ScD10A-2R | ACCTTGCGTTTCTTCTTTGGATCGCCGCCCAACTGAGAGA |
| ScD10A-3F | CCAAAGAAGAAACGCAAGGTCG |
| S-3R | CCGCCGCCAAGTGATTCTTAG |
| ScD10A-4F | CTAAGAATCACTTGGCGGCGG |
| ScD10A-4R | TCTCAACTCCTTTAAGCTTAATTAA |
| SpG-1F | TAAGCTTAAAGGAGTTGAGAATGGAT |
| SpG-1R | AAAACCACCATATTTTTTTGGATCC |
| SpG-2F | CAAAAAAATATGGTGGTTTTCTGTGGCCAACGGTAGCTTAT |
| SpG-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTACGATCAATTGTT |
| S-3F | TCTACAAAAGAAGTTTTAGATGCCAC |
| SpG-4F | CTAAGAATCACTTGGCGGC |
| SpG-4R | TCTCAACTCCTTTAAGCTTAATTAATTCT |
| SpRY-1F | AGCGGAACGCACTCGTCTCA |
| SpRY-1R | AATTGACTCCTTGGAGAATCCGC |
| SpRY-2F | GATTCTCCAAGGAGTCAATTCGCCCAAAAAGAAATTCGGACA |
| SpRY-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTTGGATCAATTGTT |
| SpRY-4F | CTAAGAATCACTTGGCGGCGGGTG |
| SpRY-4R | TGAGACGAGTGCGTTCCGCTGTCTC |
| gRNA ID | Sequence(5'-3') | PAM |
|---|---|---|
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
| Cgl17576-TCG | CACTCAGCTTCGTGCTGAAC | TCG |
| Cgl2965-TGG | ACAACCCCTGGCTCAGATGG | TGG |
| Cgl0863-CAG | TTCCACTTGGTGGACAGCAG | CAG |
| Cgl1806-CTG | CGACATAAACAAGGAGGCAC | CTG |
| Cgl2111-CCG | CGTCGAACACCTCTATCCCA | CCG |
| Cgl0179-CGG | GCACCACTGCATCTACCTGG | CGG |
| Cgl0037-GAG | TCAAGGAACATCCACCGTTC | GAG |
| Cgl2426-GTG | CACACTAGGCGCGAACTATC | GTG |
| Cgl2608-GCG | TACCACTGACCGATGCTTGC | GCG |
| Cgl0170-GGG | GTGCCTGCGATGACAAATGG | GGG |
| Cgl1467-AGA | AATCCACGTGGTAACCAGGT | AGA |
| Cgl2044-AGT | ACACATACCCCTTGCCAGAT | AGT |
| Cgl3043-AGC | CCCGAAACAAAGAGGCCATC | AGC |
| Cgl2647-TGA | CTCACAGAGTGGGCAGGCAC | TGA |
| Cgl1820-TGC | CGCTATGCGCTAGCGGTAGA | TGC |
| Cgl2711-CGA | GCCGCAGCAATTATCTCCAC | CGA |
| Cgl0698-CGT | GCACCGTGGCAGTGAGTGGC | CGT |
| Cgl2647-CGC | TCCCAGAATTACACCAGGAG | CGC |
| Cgl1890-GGA | TCCCAGCGTGCACAATACGT | GGA |
| Cgl1839-GGT | ACCACAGGTGAGACAGTTCA | GGT |
| Cgl1535-GGC | CCCCACACTTTCTCCACGAT | GGC |
| Cgl2009-AAA | CCCACCATGTCCACTATCTC | AAA |
| Cgl1008-AAT | CCCAATGTGTCCTATAGCAC | AAT |
| Cgl0170-AAC | ACCTCACCACCATCGACGAC | AAC |
| Cgl1011-TAA | CACCCCTAGAGTGAGGTGGG | TAA |
| Cgl2711-TAT | CTGGCCATTGATGCATCGGA | TAT |
| Cgl2035-TAC | ATCACACGTGTGCCGAAAAA | TAC |
| Cgl0379-CAA | AACCTCTTTGGCAACCACGA | CAA |
| Cgl2111-CAC | TCCTGCTGCTTCGCTGCTGC | CAC |
| Cgl2148-GAA | ACCAGGCACCAGCTTTCGGT | GAA |
| Cgl1108-GAT | GACCAATTTGCTTGGCCTTC | GAT |
| Cgl1239-GAC | GCACTGCATTCCGCAAACCC | GAC |
| Cgl2475-TGT | GCTCCATTCAGACCATGGAA | TGT |
| Cgl1925-CAT | CCTCTCCCAGGCACTTGTCG | CAT |
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl0982-AAG | CCAGGTTGCCGATGATTCTC | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
Table 3 gRNAs used in this study
| gRNA ID | Sequence(5'-3') | PAM |
|---|---|---|
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
| Cgl17576-TCG | CACTCAGCTTCGTGCTGAAC | TCG |
| Cgl2965-TGG | ACAACCCCTGGCTCAGATGG | TGG |
| Cgl0863-CAG | TTCCACTTGGTGGACAGCAG | CAG |
| Cgl1806-CTG | CGACATAAACAAGGAGGCAC | CTG |
| Cgl2111-CCG | CGTCGAACACCTCTATCCCA | CCG |
| Cgl0179-CGG | GCACCACTGCATCTACCTGG | CGG |
| Cgl0037-GAG | TCAAGGAACATCCACCGTTC | GAG |
| Cgl2426-GTG | CACACTAGGCGCGAACTATC | GTG |
| Cgl2608-GCG | TACCACTGACCGATGCTTGC | GCG |
| Cgl0170-GGG | GTGCCTGCGATGACAAATGG | GGG |
| Cgl1467-AGA | AATCCACGTGGTAACCAGGT | AGA |
| Cgl2044-AGT | ACACATACCCCTTGCCAGAT | AGT |
| Cgl3043-AGC | CCCGAAACAAAGAGGCCATC | AGC |
| Cgl2647-TGA | CTCACAGAGTGGGCAGGCAC | TGA |
| Cgl1820-TGC | CGCTATGCGCTAGCGGTAGA | TGC |
| Cgl2711-CGA | GCCGCAGCAATTATCTCCAC | CGA |
| Cgl0698-CGT | GCACCGTGGCAGTGAGTGGC | CGT |
| Cgl2647-CGC | TCCCAGAATTACACCAGGAG | CGC |
| Cgl1890-GGA | TCCCAGCGTGCACAATACGT | GGA |
| Cgl1839-GGT | ACCACAGGTGAGACAGTTCA | GGT |
| Cgl1535-GGC | CCCCACACTTTCTCCACGAT | GGC |
| Cgl2009-AAA | CCCACCATGTCCACTATCTC | AAA |
| Cgl1008-AAT | CCCAATGTGTCCTATAGCAC | AAT |
| Cgl0170-AAC | ACCTCACCACCATCGACGAC | AAC |
| Cgl1011-TAA | CACCCCTAGAGTGAGGTGGG | TAA |
| Cgl2711-TAT | CTGGCCATTGATGCATCGGA | TAT |
| Cgl2035-TAC | ATCACACGTGTGCCGAAAAA | TAC |
| Cgl0379-CAA | AACCTCTTTGGCAACCACGA | CAA |
| Cgl2111-CAC | TCCTGCTGCTTCGCTGCTGC | CAC |
| Cgl2148-GAA | ACCAGGCACCAGCTTTCGGT | GAA |
| Cgl1108-GAT | GACCAATTTGCTTGGCCTTC | GAT |
| Cgl1239-GAC | GCACTGCATTCCGCAAACCC | GAC |
| Cgl2475-TGT | GCTCCATTCAGACCATGGAA | TGT |
| Cgl1925-CAT | CCTCTCCCAGGCACTTGTCG | CAT |
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl0982-AAG | CCAGGTTGCCGATGATTCTC | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
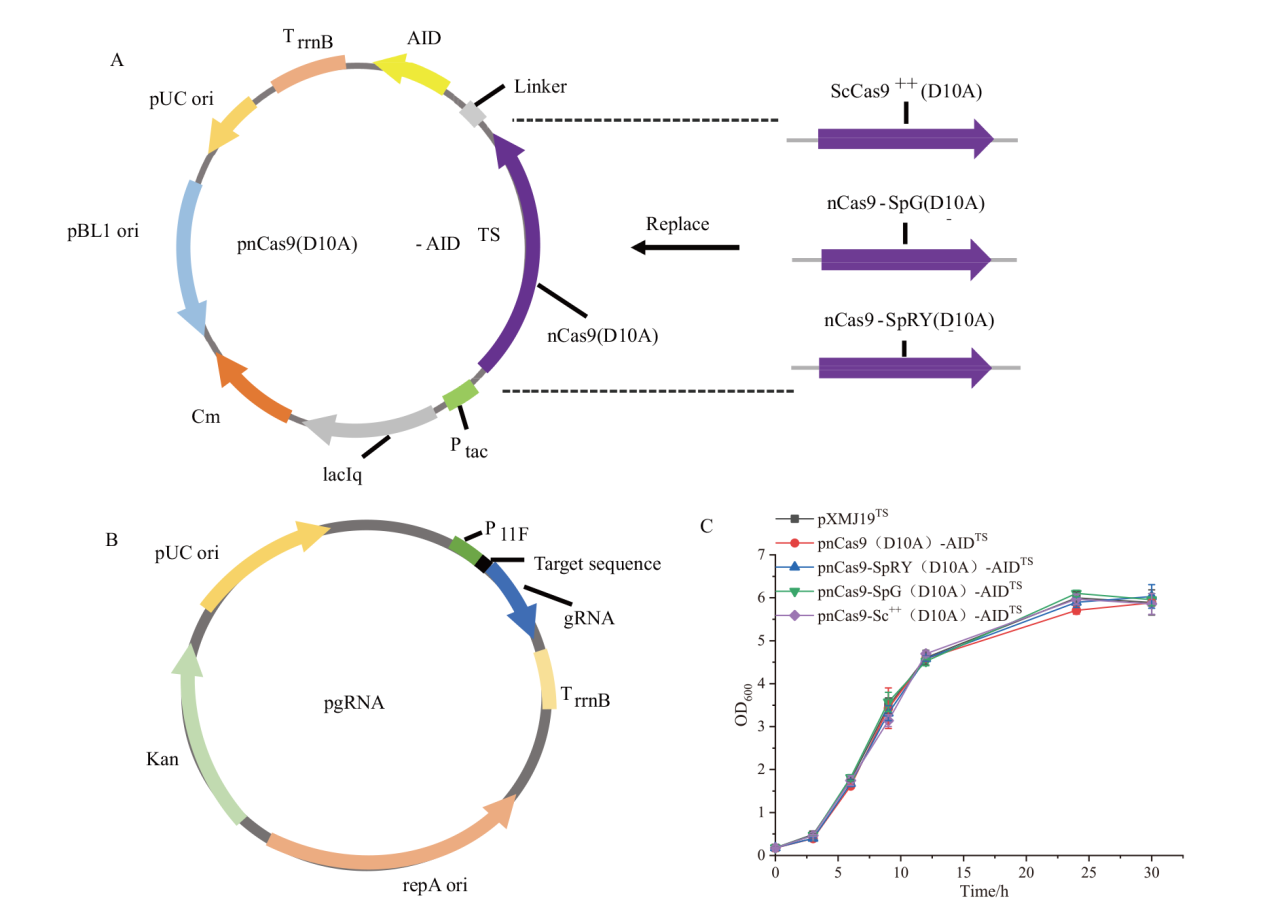
Fig. 1 Plasmid construction for base editing A: Plasmid construction for base editing with different Cas9 mutants(Ptac: Tac promoter; TrrnB: rrnB terminator; Cm: chloroamphenicol). B: Plasmid construction for gRNA expression(P11F: a derivative of cspB promoter from C. glutamicum[22]; Kan:Kanamycin). C: Effect of cytosine base editing tools combined with different Cas9 proteins on strain growth(Number of replicates n=3)

Fig. 2 Base editing results of nCas9-SpRY(D10A)-AID A: Editing efficiency for NGN PAMs. B: Editing efficiency for NAN PAMs. Number of replicates n=3, * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.000 1, student’s two-tailed t-test, the same below
| [1] |
Wang Y, Liu Y, Zheng P, et al. Microbial base editing: a powerful emerging technology for microbial genome engineering[J]. Trends Biotechnol, 2021, 39(2): 165-180.
doi: 10.1016/j.tibtech.2020.06.010 pmid: 32680590 |
| [2] |
Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424.
doi: 10.1038/nature17946 |
| [3] | Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): eaaf8729. |
| [4] |
Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471.
doi: 10.1038/nature24644 URL |
| [5] | Zhao DD, Li J, Li SW, et al. Publisher correction: Glycosylase base editors enable C-to-A and C-to-G base changes[J]. Nat Biotechnol, 2021, 39(1): 115. |
| [6] |
Deng C, Lv XQ, Li JH, et al. Development of a DNA double-strand break-free base editing tool in Corynebacterium glutamicum for genome editing and metabolic engineering[J]. Metab Eng Commun, 2020, 11: e00135.
doi: 10.1016/j.mec.2020.e00135 URL |
| [7] |
Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157.
doi: 10.1038/s41586-019-1711-4 |
| [8] |
Tong YJ, Jørgensen TS, Whitford CM, et al. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing[J]. Nat Commun, 2021, 12: 5206.
doi: 10.1038/s41467-021-25541-3 |
| [9] |
Yang B, Yang L, Chen J. Development and application of base editors[J]. CRISPR J, 2019, 2(2): 91-104.
doi: 10.1089/crispr.2019.0001 pmid: 30998092 |
| [10] |
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nat Biotechnol, 2020, 38(7): 824-844.
doi: 10.1038/s41587-020-0561-9 pmid: 32572269 |
| [11] |
Yu SY, Birkenshaw A, Thomson T, et al. Increasing the targeting scope of CRISPR base editing system beyond NGG[J]. CRISPR J, 2022, 5(2): 187-202.
doi: 10.1089/crispr.2021.0109 URL |
| [12] |
Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296.
doi: 10.1126/science.aba8853 pmid: 32217751 |
| [13] |
Chatterjee P, Jakimo N, Lee J, et al. An engineered ScCas9 with broad PAM range and high specificity and activity[J]. Nat Biotechnol, 2020, 38(10): 1154-1158.
doi: 10.1038/s41587-020-0517-0 |
| [14] |
Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels-Corynebacterium glutamicum as versatile cell factory[J]. Curr Opin Biotechnol, 2012, 23(4): 631-640.
doi: 10.1016/j.copbio.2011.11.012 URL |
| [15] |
Wang Y, Liu Y, Liu J, et al. MACBETH: Multiplex automated Corynebacterium glutamicum base editing method[J]. Metab Eng, 2018, 47: 200-210.
doi: S1096-7176(17)30417-2 pmid: 29580925 |
| [16] |
Wang Y, Liu Y, Li JW, et al. Expanding targeting scope, editing window, and base transition capability of base editing in Corynebacterium glutamicum[J]. Biotechnol Bioeng, 2019, 116(11): 3016-3029.
doi: 10.1002/bit.27121 pmid: 31317533 |
| [17] |
黄华媚, 白立宽, 刘叶, 等. BE3型胞嘧啶碱基编辑器在谷氨酸棒杆菌中的开发及应用[J]. 生物技术通报, 2020, 36(3): 95-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0956 |
|
Huang HM, Bai LK, Liu Y, et al. Development and application of BE3 cytidine base editor in Corynebacterium glutamicum[J]. Biotechnol Bull, 2020, 36(3): 95-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0956 |
|
| [18] | 李俊维, 刘叶, 王钰, 等. 谷氨酸棒杆菌碱基编辑的条件优化[J]. 生物工程学报, 2020, 36(1): 143-151. |
| Li JW, Liu Y, Wang Y, et al. Optimization of base editing in Cory-nebacterium glutamicum[J]. Chin J Biotechnol, 2020, 36(1): 143-151. | |
| [19] | 卢挥, 张启, 于思礼, 等. 谷氨酸棒杆菌中基于CRISPR/Cas9的多位点碱基编辑系统的优化[J]. 生物工程学报, 2022, 38(2): 780-795. |
| Lu H, Zhang Q, Yu SL, et al. Optimization of CRISPR/Cas9-based multiplex base editing in Corynebacterium glutamicum[J]. Chin J Biotechnol, 2022, 38(2): 780-795. | |
| [20] |
Ruan YL, Zhu LJ, Li Q. Improving the electro-transformation efficiency of Corynebacterium glutamicum by weakening its cell wall and increasing the cytoplasmic membrane fluidity[J]. Biotechnol Lett, 2015, 37(12): 2445-2452.
doi: 10.1007/s10529-015-1934-x URL |
| [21] |
Kluesner MG, Nedveck DA, Lahr WS, et al. EditR: a method to quantify base editing from Sanger sequencing[J]. CRISPR J, 2018, 1(3): 239-250.
doi: 10.1089/crispr.2018.0014 URL |
| [22] |
Peyret JL, Bayan N, Joliff G, et al. Characterization of the cspB gene encoding PS2, an ordered surface-layer protein in Corynebacterium glutamicum[J]. Mol Microbiol, 1993, 9(1): 97-109.
doi: 10.1111/j.1365-2958.1993.tb01672.x pmid: 8412676 |
| [23] |
Wang Y, Cheng HJ, Liu Y, et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing[J]. Nat Commun, 2021, 12: 678.
doi: 10.1038/s41467-021-21003-y pmid: 33514753 |
| [24] | Liu Y, Wang RY, Liu JH, et al. Base editor enables rational genome-scale functional screening for enhanced industrial phenotypes in Corynebacterium glutamicum[J]. Sci Adv, 2022, 8(35): eabq2157. |
| [25] |
Tian KR, Hong X, Guo MM, et al. Development of base editors for simultaneously editing multiple loci in Lactococcus lactis[J]. ACS Synth Biol, 2022, 11(11): 3644-3656.
doi: 10.1021/acssynbio.1c00561 URL |
| [1] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [2] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [3] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [4] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [5] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [6] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [7] | LU Zhen-wan, LI Xue-qi, HUANG Jin-guang, ZHOU Huan-bin. Creation of Glyphosate-tolerant Rice by Cytosine Base Editing [J]. Biotechnology Bulletin, 2023, 39(2): 63-69. |
| [8] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [9] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [10] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [11] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [12] | TANG Guang-fu, GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong. Editing pyrG Gene of Monascus by CRISPR/Cas 9 and Its Effects on Secondary Metabolism [J]. Biotechnology Bulletin, 2022, 38(8): 198-205. |
| [13] | LAI Xin-tong, WANG Ke-lan, YOU Yu-xin, TAN Jun-jie. Recent Advances in CRISPR/Cas-based DNA Base Editing [J]. Biotechnology Bulletin, 2022, 38(6): 1-12. |
| [14] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [15] | CHEN Ying-dan, ZHANG Yang, XIA Qiang, SUN Hong-xia. Gene Editing Technology of CRISPR/Cas and Its Applications in Microalgae Research [J]. Biotechnology Bulletin, 2022, 38(5): 257-268. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||