








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (3): 181-192.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0960
Previous Articles Next Articles
MEI Xian-jun1,2( ), SONG Hui-yang2, LI Jing-hao2, MEI Chao2,3, SONG Qian-na2,3, FENG Rui-yun2,3(
), SONG Hui-yang2, LI Jing-hao2, MEI Chao2,3, SONG Qian-na2,3, FENG Rui-yun2,3( ), CHEN Xi-ming1,2(
), CHEN Xi-ming1,2( )
)
Received:2023-10-11
Online:2024-03-26
Published:2024-04-08
Contact:
FENG Rui-yun, CHEN Xi-ming
E-mail:1732244325@qq.com;516834898@qq.com;fengruiyun1970@163.com
MEI Xian-jun, SONG Hui-yang, LI Jing-hao, MEI Chao, SONG Qian-na, FENG Rui-yun, CHEN Xi-ming. Cloning and Expression Analysis of StDof5 Gene in Potato[J]. Biotechnology Bulletin, 2024, 40(3): 181-192.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| Actin-F | GGGATGGAGAAGTTTGGTGGTGG |
| Actin-R | CTTCGACCAAGGGATGGTGTAGC |
| Dof-F.2 | GCTTGACTTTGATGGAGTGGTGG |
| Dof-R.2 | AAGATTGGCTATCGGAGGTGC |
| ABF4-F | CTTCTGCACATACAAGGATATTCAA |
| ABF4-R | TGCCTAGCCAAAGGGAAATCA |
| MYB2-F | ATGACATGGACCTCCGACGA |
| MYB2-R | CGTTCGCTTTAGACCAGCAC |
| SnRK2s-F | TTGCTGTAGAGAAGGTGGGT |
| SnRK2s-R | GTCCTACTGTCACTGCCGTC |
| DREB2A-F | CACAGAAGAAAAGAGAAATGGCT |
| DREB2A-R | TCCACTATAATCCAACGGCAGA |
| ABI3-F | TGCTGACATGAAGTGTGGCA |
| ABI3-R | TTTTGCCCTCCGACTTTGGT |
| ABI5-F | TAAAACAGGCCCTGGCGGAG |
| ABI5-R | TTGCGCCTTTGTTTGAGCTT |
Table 1 Primer sequences for fluorescence quantification
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| Actin-F | GGGATGGAGAAGTTTGGTGGTGG |
| Actin-R | CTTCGACCAAGGGATGGTGTAGC |
| Dof-F.2 | GCTTGACTTTGATGGAGTGGTGG |
| Dof-R.2 | AAGATTGGCTATCGGAGGTGC |
| ABF4-F | CTTCTGCACATACAAGGATATTCAA |
| ABF4-R | TGCCTAGCCAAAGGGAAATCA |
| MYB2-F | ATGACATGGACCTCCGACGA |
| MYB2-R | CGTTCGCTTTAGACCAGCAC |
| SnRK2s-F | TTGCTGTAGAGAAGGTGGGT |
| SnRK2s-R | GTCCTACTGTCACTGCCGTC |
| DREB2A-F | CACAGAAGAAAAGAGAAATGGCT |
| DREB2A-R | TCCACTATAATCCAACGGCAGA |
| ABI3-F | TGCTGACATGAAGTGTGGCA |
| ABI3-R | TTTTGCCCTCCGACTTTGGT |
| ABI5-F | TAAAACAGGCCCTGGCGGAG |
| ABI5-R | TTGCGCCTTTGTTTGAGCTT |

Fig. 1 Bioinformatics analysis of StDof5 protein A: Transmembrane domain of the StDof5 protein; B: secondary structure prediction of potato StDof5 protein; C: hydrophilic nature of potato StDof5 protein; D: tertiary structure prediction of potato StDof5 protein
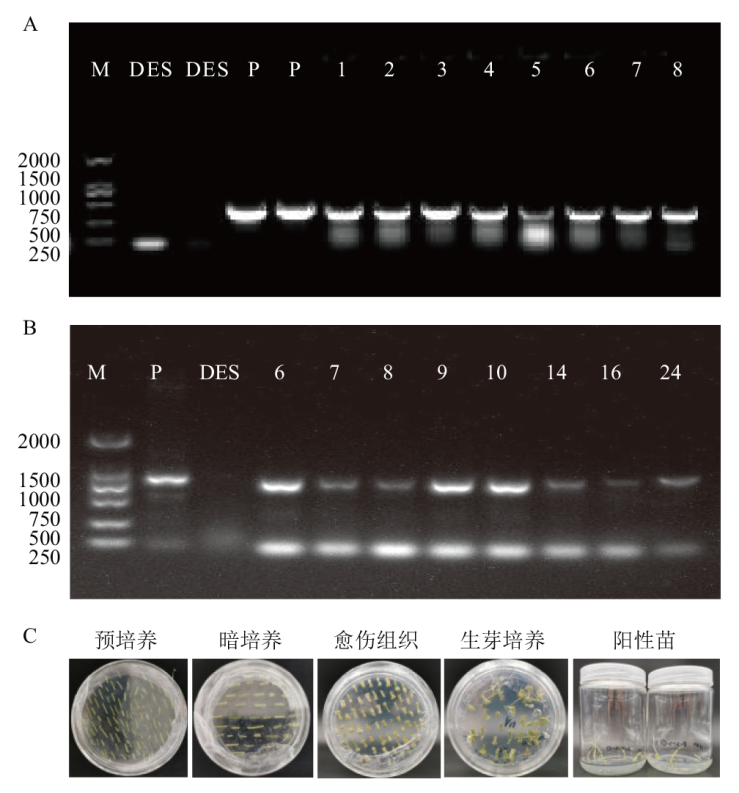
Fig. 5 Construction of the potato StDof5 vector A: Agrobacterium vector construction; B: potato genetically transformed tissue culture; C: identification of overexpressed potato plants. M: Marker 2000 bp. P: Plasmid. DES: Wild type. 1-8: Positive single clone. 6-10, 14, 16, 24: Overexpression positive plants
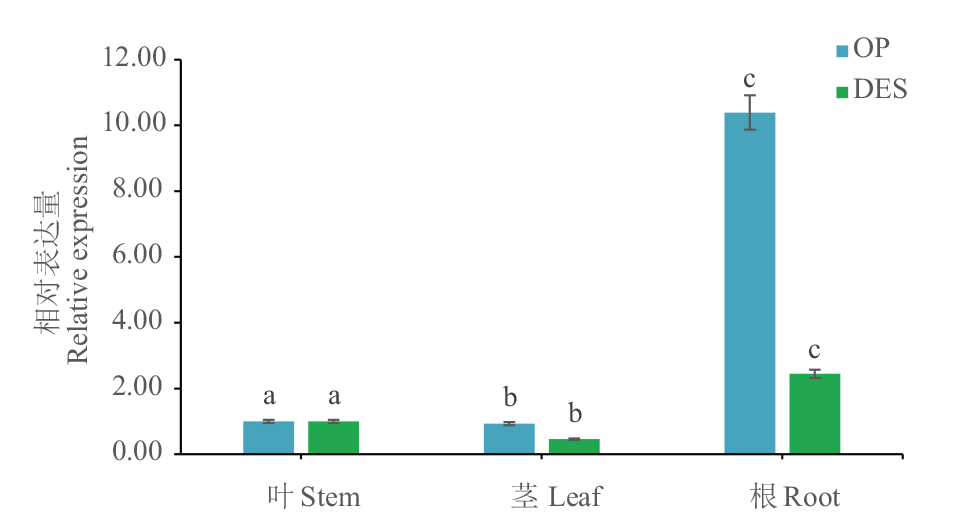
Fig. 6 Analysis of Dof5 gene expression in different tissues of potato DES: Wild type. OP: Overexpressed positive plants. Different letters indicate a significant difference(P<0.05). The same below
| 测量指标Measurement index | 盐浓度Salting strength/(mol·L-1) | 品种名称Variety name | ||||||
|---|---|---|---|---|---|---|---|---|
| DES | OP-6 | OP-7 | OP-9 | OP-10 | OP-16 | OP-24 | ||
| 株高Plant height/cm | 0 | 6.87±0.38a | 7.10±0.17a | 5.57±0.40a | 5.43±0.15a | 6.53±0.85a | 7.37±0.75a | 7.83±1.01a |
| 50 | 3.70±010b | 4.83±0.65b | 3.70±0.36b | 4.37±0.21b | 3.97±0.35b | 6.00±0.72b | 4.13±0.45b | |
| 100 | 2.13±0.76c | 2.97±0.42c | 2.00±0.53c | 1.77±0.31c | 2.80±0.36c | 3.00±0.44c | 2.93±0.15c | |
| 鲜重Fresh weight/g | 0 | 0.21±0.01a | 0.23±0.01a | 0.19±0.01a | 0.21±0.02a | 0.24±0.02a | 0.19±0.02a | 0.35±0.01a |
| 50 | 0.14±0.01b | 0.19±0.02b | 0.12±0.02b | 0.16±0.02b | 0.16±0.02b | 0.16±0.01a | 0.28±0.02b | |
| 100 | 0.09±0.01c | 0.12±0.02c | 0.09±0.01c | 0.09±0.01c | 0.12±0.01c | 0.12±0.02b | 0.15±0.02c | |
| 根长Root length/cm | 0 | 12.13±0.25a | 12.20±0.46a | 12.23±2.03a | 14.70±0.62a | 17.63±0.35a | 12.90±1.49a | 12.03±0.72a |
| 50 | 9.00±0.26b | 9.73±0.12b | 10.93±1.79ab | 13.07±0.31b | 13.73±0.31b | 10.53±0.61b | 11.67±0.31a | |
| 100 | 7.53±0.25c | 8.93±060b | 6.70±2.74b | 4.00±0.72c | 10.1±0.20c | 9.73±0.31b | 8.60±0.53b | |
| 根数Root | 0 | 6.33±1.15a | 7.33 ±2.31a | 8.00±0.00a | 7.00±1.00a | 8.67±1.53a | 6.00±0.00a | 9.67±2.08a |
| 50 | 3.67±2.08a | 5.67±2.31a | 4.00±1.00b | 5.33±2.31a | 4.67±1.53b | 6.67±2.89a | 5.67±1.15b | |
| 100 | 3.33±1.53a | 5.00±1.00a | 3.33±2.08b | 1.00±0.00b | 4.33±2.31b | 1.67±0.58b | 3.00±1.73b | |
| 叶片数Number of leaves | 0 | 7.33±0.58a | 8.33±0.58a | 7.67±0.58a | 6.67±0.58a | 7.00±0.00a | 7.00±1.00a | 6.67±0.58a |
| 50 | 6.33±0.58a | 7.00±0.00a | 6.33±1.15a | 6.00±1.00ab | 7.00±0.00a | 7.00±1.00a | 6.66±0.58a | |
| 100 | 7.00±1.00a | 7.00±1.00a | 6.67±0.58a | 5.00±0.00b | 6.33±1.15a | 6.00±1.00a | 6.67±0.58a | |
Table 2 Determination of the growth indicators of salt stress
| 测量指标Measurement index | 盐浓度Salting strength/(mol·L-1) | 品种名称Variety name | ||||||
|---|---|---|---|---|---|---|---|---|
| DES | OP-6 | OP-7 | OP-9 | OP-10 | OP-16 | OP-24 | ||
| 株高Plant height/cm | 0 | 6.87±0.38a | 7.10±0.17a | 5.57±0.40a | 5.43±0.15a | 6.53±0.85a | 7.37±0.75a | 7.83±1.01a |
| 50 | 3.70±010b | 4.83±0.65b | 3.70±0.36b | 4.37±0.21b | 3.97±0.35b | 6.00±0.72b | 4.13±0.45b | |
| 100 | 2.13±0.76c | 2.97±0.42c | 2.00±0.53c | 1.77±0.31c | 2.80±0.36c | 3.00±0.44c | 2.93±0.15c | |
| 鲜重Fresh weight/g | 0 | 0.21±0.01a | 0.23±0.01a | 0.19±0.01a | 0.21±0.02a | 0.24±0.02a | 0.19±0.02a | 0.35±0.01a |
| 50 | 0.14±0.01b | 0.19±0.02b | 0.12±0.02b | 0.16±0.02b | 0.16±0.02b | 0.16±0.01a | 0.28±0.02b | |
| 100 | 0.09±0.01c | 0.12±0.02c | 0.09±0.01c | 0.09±0.01c | 0.12±0.01c | 0.12±0.02b | 0.15±0.02c | |
| 根长Root length/cm | 0 | 12.13±0.25a | 12.20±0.46a | 12.23±2.03a | 14.70±0.62a | 17.63±0.35a | 12.90±1.49a | 12.03±0.72a |
| 50 | 9.00±0.26b | 9.73±0.12b | 10.93±1.79ab | 13.07±0.31b | 13.73±0.31b | 10.53±0.61b | 11.67±0.31a | |
| 100 | 7.53±0.25c | 8.93±060b | 6.70±2.74b | 4.00±0.72c | 10.1±0.20c | 9.73±0.31b | 8.60±0.53b | |
| 根数Root | 0 | 6.33±1.15a | 7.33 ±2.31a | 8.00±0.00a | 7.00±1.00a | 8.67±1.53a | 6.00±0.00a | 9.67±2.08a |
| 50 | 3.67±2.08a | 5.67±2.31a | 4.00±1.00b | 5.33±2.31a | 4.67±1.53b | 6.67±2.89a | 5.67±1.15b | |
| 100 | 3.33±1.53a | 5.00±1.00a | 3.33±2.08b | 1.00±0.00b | 4.33±2.31b | 1.67±0.58b | 3.00±1.73b | |
| 叶片数Number of leaves | 0 | 7.33±0.58a | 8.33±0.58a | 7.67±0.58a | 6.67±0.58a | 7.00±0.00a | 7.00±1.00a | 6.67±0.58a |
| 50 | 6.33±0.58a | 7.00±0.00a | 6.33±1.15a | 6.00±1.00ab | 7.00±0.00a | 7.00±1.00a | 6.66±0.58a | |
| 100 | 7.00±1.00a | 7.00±1.00a | 6.67±0.58a | 5.00±0.00b | 6.33±1.15a | 6.00±1.00a | 6.67±0.58a | |

Fig. 9 Growth status of tissue culture seedlings after 25 d of salt stress From top to bottom, the pictures are: DES, OP-6, OP-7, OP-9, OP-10, OP-16, OP-24, with the salt stress of 0, 50 and 100 mmol/L NaCl, respectively
| [1] |
Gupta S, Malviya N, Kushwaha H, et al. Insights into structural and functional diversity of Dof(DNA binding with one finger)transcription factor[J]. Planta, 2015, 241(3): 549-562.
doi: 10.1007/s00425-014-2239-3 pmid: 25564353 |
| [2] |
Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone[J]. Plant Physiol, 2003, 133(2): 850-863.
pmid: 14500792 |
| [3] |
Wang CM, Song BB, Dai YQ, et al. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear(Pyrus bretschneideri)[J]. BMC Plant Biol, 2021, 21(1): 235.
doi: 10.1186/s12870-021-03024-3 |
| [4] |
Wang T, Yue JJ, Wang XJ, et al. Genome-wide identification and characterization of the Dof gene family in moso bamboo(Phyllostachys heterocycla var. pubescens)[J]. Genes Genom, 2016, 38(8): 733-745.
doi: 10.1007/s13258-016-0418-2 URL |
| [5] | Li Y, Ding WJ, Jiang HY, et al. Advances in research of the Dof gene family in plant[J]. Acta Botanica Boreali-Occidentalia Sinica, 2018, 38(9): 1758-1766. |
| [6] |
Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors[J]. Plant J, 1999, 17(2): 209-214.
doi: 10.1046/j.1365-313x.1999.00363.x pmid: 10074718 |
| [7] |
Yanagisawa S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants[J]. Plant Cell Physiol, 2004, 45(4): 386-391.
doi: 10.1093/pcp/pch055 pmid: 15111712 |
| [8] |
Venkatesh J, Park SW. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger(Dof)transcription factor family in potato[J]. Plant Physiol Biochem, 2015, 94: 73-85.
doi: 10.1016/j.plaphy.2015.05.010 URL |
| [9] |
Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif[J]. J Biol Chem, 1993, 268(21): 16028-16036.
pmid: 8340424 |
| [10] | 张华珍, 吴昊, 李香花, 等. 一个低氮诱导表达的水稻Dof转录因子OsDof-13的分离和转化[J]. 分子植物育种, 2007, 5(4): 455-460. |
| Zhang HZ, Wu H, Li XH, et al. Isolation and transformation of OsDof-13, a member of rice Dof transcription factor family, induced by low nitrogen stress[J]. Mol Plant Breed, 2007, 5(4): 455-460. | |
| [11] |
Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families[J]. BMC Evol Biol, 2003, 3: 17.
pmid: 12877745 |
| [12] |
Cai XF, Zhang YY, Zhang CJ, et al. Genome-wide analysis of plant-specific Dof transcription factor family in tomato[J]. J Integr Plant Biol, 2013, 55(6): 552-566.
doi: 10.1111/jipb.v55.6 URL |
| [13] | 马凯恒, 张彤, 闫鹏宇, 等. 核桃转录因子JrDof3的克隆及渗透胁迫表达分析[J]. 中南林业科技大学学报, 2020, 40(10): 35-41. |
| Ma KH, Zhang T, Yan PY, et al. Cloning and expression analysis of walnut transcription factor Dof3 to osmotic stress[J]. J Central South Univ For Technol, 2020, 40(10): 35-41. | |
| [14] |
Corrales AR, Nebauer SG, Carrillo L, et al. Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses[J]. J Exp Bot, 2014, 65(4): 995-1012.
doi: 10.1093/jxb/ert451 URL |
| [15] | 成亮, 刘宝玲, 刘盼, 等. 青狗尾草Dof基因家族的全基因组鉴定及组织特异性表达分析[J]. 分子植物育种, 2018, 16(13): 4226-4234. |
| Cheng L, Liu BL, Liu P, et al. Genome-wide analysis and expression profiling of dof family in Setaria viridis[J]. Mol Plant Breed, 2018, 16(13): 4226-4234. | |
| [16] |
Choi HI, Hong JH, Ha JO, et al. ABFs, a family of ABA-responsive element binding factors[J]. J Biol Chem, 2000, 275(3): 1723-1730.
doi: 10.1074/jbc.275.3.1723 pmid: 10636868 |
| [17] | 李杨. 玉米中PP2Cs与SnRK2s相互作用研究[D]. 合肥: 安徽农业大学, 2021. |
| Li Y. Study on the interaction between PP2Cs and SnRK2s in maize[D]. Hefei: Anhui Agricultural University, 2021. | |
| [18] | 赵芝婧. 拟南芥转录因子LFY2和MYB2在花药发育调控网络中的作用研究[D]. 北京林业大学, 2020. |
| Zhao ZJ. Role of Arabidopsis transcription factors LFY2 and MYB2 in anther development regulation network[D]. Beijing: Beijing Forestry University, 2020. | |
| [19] |
Achard P, Gong F, Cheminant S, et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism[J]. Plant cell, 2008, 20(8): 2117-2129.
doi: 10.1105/tpc.108.058941 pmid: 18757556 |
| [20] |
Fujita Y, Fujita AM, Satoh CR, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis[J]. Plant Cell, 2005, 17(12): 3470-3488.
doi: 10.1105/tpc.105.035659 URL |
| [21] |
Zheng ZF, Xu XP, Crosley RA, et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis[J]. Plant Physiol, 2010, 153(1): 99-113.
doi: 10.1104/pp.109.150789 URL |
| [22] |
Wang YP, Li L, Ye TT, et al. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis[J]. J Exp Bot, 2013, 64(2): 675-684.
doi: 10.1093/jxb/ers361 URL |
| [23] | 王伟, 李立芹, 邹雪, 等. 马铃薯块茎总RNA提取方法比较[J]. 江苏农业科学, 2013, 41(1): 38-40. |
| Wang W, Li LQ, Zou X, et al. Comparison of extraction methods of total RNA from potato tuber[J]. Jiangsu Agric Sci, 2013, 41(1): 38-40. | |
| [24] | 于惠琳, 吴玉群, 王延波, 等. 甜玉米籽粒总RNA提取方法的比较[J]. 园艺与种苗, 2022, 42(1): 67-68, 71. |
| Yu HL, Wu YQ, Wang YB, et al. Comparison of total RNA extraction methods from sweet corn kernels[J]. Hortic Seed, 2022, 42(1): 67-68, 71. | |
| [25] | 张晓丽, 代红军. 植物RNA提取方法的研究进展[J]. 北方园艺, 2014(8): 175-178. |
| Zhang XL, Dai HJ. Research progress on extraction method of plant RNA[J]. North Hortic, 2014(8): 175-178. | |
| [26] | 赵勤. 农杆菌介导的马铃薯遗传转化影响因素研究进展[J]. 园艺与种苗, 2011, 31(2): 92-94. |
| Zhao Q. Study on influencing factors of Agrobacterium-mediated genetic transformation in potato[J]. Rain Fed Crops, 2011, 31(2): 92-94. | |
| [27] | 杨永智. 农杆菌介导法马铃薯遗传转化体系的优化[J]. 江苏农业学报, 2013, 29(4): 738-742. |
| Yang YZ. Optimization of Agrobacterium tumerfaciens mediated potato transformation system[J]. Jiangsu J Agric Sci, 2013, 29(4): 738-742. | |
| [28] | 连欢欢, 杨永智, 王舰. 根癌农杆菌介导的马铃薯DM茎段遗传转化条件的优化[J]. 分子植物育种, 2016, 14(6): 1491-1499. |
| Lian HH, Yang YZ, Wang J. Optimization of conditions for genetic transformation of potato DM stems[J]. Molecular plant breeding, 2016, 14(6): 1491-1499. | |
| [29] | 张萍. 根癌农杆菌介导的马铃薯遗传转化体系的研究[J]. 陕西农业科学, 2019, 65(4): 27-29, 40. |
| Zhang P. Study on genetic transformation system of potato mediated by Agrobacterium tumefaciens[J]. Shaanxi J Agric Sci, 2019, 65(4): 27-29, 40. | |
| [30] | 李逸菲, 王瑞博, 夏惠婷, 等. 农杆菌介导四季秋海棠遗传转化体系建立[J]. 东北农业大学学报, 2023, 54(5): 19-27. |
| Li YF, Wang RB, Xia HT, et al. Establishment of Agrobacteri-um-mediated genetic transformation system for Begonia semperflo-rens[J]. J Northeast Agric Univ, 2023, 54(5): 19-27. | |
| [31] | 宋倩娜, 梅超, 霍利光, 等. 马铃薯品种‘并薯6号’遗传转化体系的建立[J]. 中国马铃薯, 2021, 35(5): 385-396. |
| Song QN, Mei C, Huo LG, et al. Establishment of the genetic transformation system for potato variety ‘Bingshu 6’[J]. Chin potato J, 2021, 35(5): 385-396. | |
| [32] | 高志鹏, 杜玉萧, 冯霞, 等. 鄂马铃薯3号遗传转化体系的构建与表达[J]. 湖北农业科学, 2022, 61(9): 162-167, 191. |
| Gao ZP, Du YX, Feng X, et al. Construction and expression of the # 3 genetic transformation system of Hubei potato No.3[J]. Hubei Agric Sci, 2022, 61(9): 162-167, 191. | |
| [33] | 陈兆贵, 林燕文, 李希陶, 等. 马铃薯植株再生和遗传转化技术研究进展[J]. 智慧农业导刊, 2023, 3(9): 12-15. |
| Chen ZG, Lin YW, Li XT, et al. Research progress of plant regeneration and genetic transformation of potatoes[J]. J Smart Agri, 2023, 3(9): 12-15. | |
| [34] |
公鑫, 王业红, 张剑峰, 等. 马铃薯RPP13基因RNAi载体的构建及遗传转化[J]. 华北农学报, 2023, 38(1): 178-183.
doi: 10.7668/hbnxb.20193453 |
| Gong X, Wang YH, Zhang JF, et al. Construction and genetic transformation of the RNAi vector of potato RPP 13 gene[J]. Acta Agric Boreali Sin, 2023, 38(1): 178-183. | |
| [35] | 万丽丽, 王转茸, 汤谧, 等. 甜瓜遗传转化技术的优化及DRs类基因的应用[J]. 江苏农业科学, 2023, 51(7): 49-58. |
| Wan LL, Wang ZR, Tang M, et al. Optimization of genetic transformation technology of melon and the application of DRs class genes[J]. Jiangsu Agric Sci, 2023, 51(7): 49-58. | |
| [36] |
Corrales AR, Carrillo L, Lasierra P, et al. Multifaceted role of cycling DOF factor 3(CDF3)in the regulation of flowering time and abiotic stress responses in Arabidopsis[J]. Plant Cell Environ, 2017, 40(5): 748-764.
doi: 10.1111/pce.v40.5 URL |
| [37] |
Xu PP, Chen HY, Ying L, et al. AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana[J]. Sci Rep, 2016, 6: 27705.
doi: 10.1038/srep27705 |
| [38] |
van Zelm E, Zhang YX, Testerink C. Salt tolerance mechanisms of plants[J]. Annu Rev Plant Bio, 2020, 71: 403-433.
doi: 10.1146/annurev-arplant-050718-100005 URL |
| [39] |
Su Y, Liang W, Liu ZJ, et al. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum[J]. J Plant Physiol, 2017, 218: 222-234.
doi: 10.1016/j.jplph.2017.07.017 URL |
| [40] |
Cai XF, Zhang CJ, Shu WB, et al. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato[J]. Biochem Biophys Res Commun, 2016, 474(4): 736-741.
doi: 10.1016/j.bbrc.2016.04.148 URL |
| [41] |
Kim J, Kim JH, Lyu JI, et al. New insights into the regulation of leaf senescence in Arabidopsis[J]. J Exp Bot, 2018, 69(4): 787-799.
doi: 10.1093/jxb/erx287 URL |
| [42] |
Lee BD, Ri KM, Young KM, et al. The F-box protein FKF1inhibits dimerization of COP1 in the control of photoperiodic flowering[J]. Nature Communications, 2018, 8(1): 2259.
doi: 10.1038/s41467-017-02476-2 |
| [43] |
Wen CL, Cheng Q, Zhao LQ, et al. Identification and characterisation of Dof transcription factors in the cucumber genome[J]. Sci Rep, 2016, 6: 23072.
doi: 10.1038/srep23072 |
| [44] |
Noguero M, Atif RM, OCHATT S, et al. The role of the DNA-binding One Zinc Finger(Dof)transcription factor family in plants[J]. Plant Sci, 2013, 209: 32-45.
doi: 10.1016/j.plantsci.2013.03.016 URL |
| [45] |
Zhang ZR, Yuan L, Liu X, et al. Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica[J]. Gene, 2018, 639: 137-148.
doi: 10.1016/j.gene.2017.09.039 URL |
| [46] |
Ewas M, Khames E, et al. The tomato Dof Daily Fluctuations 1, DESDF1 acts as flowering accelerator and protector against various stresses[J]. Sci Rep, 2017, 7(1): 10299.
doi: 10.1038/s41598-017-10399-7 |
| [47] |
Ma J, Li MY, Wang F, et al. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage[J]. BMC Genomics, 2015, 16(1): 33.
doi: 10.1186/s12864-015-1242-9 URL |
| [48] |
Kang Y, Yang X, Liu Y, et al. Integration of mRNA and miRNA analysis reveals the molecular mechanism of potato(Solanum tuberosum L.)response to alkali stress[J]. International Journal of Biological Macromolecules, 2021, 182: 938-949.
doi: 10.1016/j.ijbiomac.2021.04.094 URL |
| [1] | LIU Wen-jin, MA Rui, LIU Sheng-yan, YANG Jiang-wei, ZHANG Ning, SI Huai-jun. Cloning of StCIPK11 Gene and Analysis of Its Response to Drought Stress in Solanum tuberosum [J]. Biotechnology Bulletin, 2023, 39(9): 147-155. |
| [2] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [3] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [4] | AN Miao, WANG Tong-tong, FU Yi-ting, XIA Jun-jun, PENG Suo-tang, DUAN Yong-hong. Genetic Diversity Analysis and Molecular Identity Card Construction by SSR Markers of 52 Solanum tuberosum L. Varieties(Lines) [J]. Biotechnology Bulletin, 2023, 39(12): 136-147. |
| [5] | TAO Na, LI Mao-xing, GUO Hua-chun. Optimization of Sweet Potato Genetic Transformation System Mediated by Agrobacterium rhizogenes [J]. Biotechnology Bulletin, 2023, 39(10): 175-183. |
| [6] | ZHOU Jia-yan, ZOU Jian, CHEN Wei-ying, WU Yi-chao, CHEN Xi-tong, WANG Qian, ZENG Wen-jing, HU Nan, YANG Jun. Construction of Multi-gene Interference System for Plant and Analysis of Its Application Efficiency [J]. Biotechnology Bulletin, 2023, 39(1): 115-126. |
| [7] | SUN Wei, ZHANG Yan, WANG Yu-han, XU Hui, XU Xiao-rong, JU Zhi-gang. Cloning of Rd3GT1 in Rhododendron delavayi and Its Effect on Flower Color Formation of Petunia hybrida [J]. Biotechnology Bulletin, 2022, 38(9): 198-206. |
| [8] | YU Guo-hong, LIU Peng-cheng, LI Lei, LI Ming-zhe, CUI Hai-ying, HAO Hong-bo, GUO An-qiang. Physiological Responses of Potato in Different Genotypes to Drought Stress [J]. Biotechnology Bulletin, 2022, 38(5): 56-63. |
| [9] | YANG Jia-hui, SUN Yu-ping, LU Ya-ning, LIU huan, LU Cun-fu, CHEN Yu-zhen. Abiotic Stress Resistance of Escherichia coli Transformed with Arabidopsis thaliana AtTERT Gene [J]. Biotechnology Bulletin, 2022, 38(2): 1-9. |
| [10] | LIU Meng-meng, HAN Li-jun, LIU Bao-ling, XUE Jin-ai, LI Run-zhi. Cloning and Expression Analysis of GhSDP1 and Its Promoter in Gossypium hirsutum [J]. Biotechnology Bulletin, 2022, 38(2): 34-43. |
| [11] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [12] | HU Zi-yao, DAI Pei-hong, LIU Chao, Madina Mulati, WANG Qian, Wugalihan Abuduwili, ZHAO Yi, SUN Ling, XU Shi-jia, LI Yue. Molecular Cloning,Expression and VIGS Construction of a Small GTP-binding Protein Gene GhROP3 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(9): 106-113. |
| [13] | GAO Bo, MA Juan, LI Xiu-hua, LI Jiao-sheng, WANG Rong-yan, CHEN Shu-long. Cloning and Functional Analysis of Dd-mel-26 Gene from Ditylenchus destructor [J]. Biotechnology Bulletin, 2021, 37(7): 107-117. |
| [14] | CUI Xiang-hua, TAO Nan, CHENG Bo-pu, ZHAO Yong-chang, CHEN Wei-min, LI Jing. Screening Promoters for Genetic Transformation of Cyclocybe aegerita [J]. Biotechnology Bulletin, 2021, 37(5): 259-266. |
| [15] | CHEN Ying, CHEN Xi, WANG Qian, WANG Xiao-li. Cloning,Expression and Biological Function Analysis of Universal Stress Protein in Festuca arundinacea [J]. Biotechnology Bulletin, 2021, 37(2): 32-39. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||