








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 141-152.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1096
Previous Articles Next Articles
LIU Bao-cai1,2( ), ZHANG Wu-jun1,2, ZHAO Yun-qing1,2, HUANG Ying-zhen1,2, CHEN Jing-ying1,2(
), ZHANG Wu-jun1,2, ZHAO Yun-qing1,2, HUANG Ying-zhen1,2, CHEN Jing-ying1,2( )
)
Received:2023-11-21
Online:2024-05-26
Published:2024-06-13
Contact:
CHEN Jing-ying
E-mail:626813844@qq.com;cjy6601@163.com
LIU Bao-cai, ZHANG Wu-jun, ZHAO Yun-qing, HUANG Ying-zhen, CHEN Jing-ying. Identification and Tissue Expression of Four Transcription Factors in Pholidota chinensis[J]. Biotechnology Bulletin, 2024, 40(5): 141-152.

Fig. 2 Characteristics of C2H2 gene family in P. chinensis A: Conserved motifs in C2H2 family; B: differential expressions of C2H2 family in different tissues; C: C2H2 family evolution
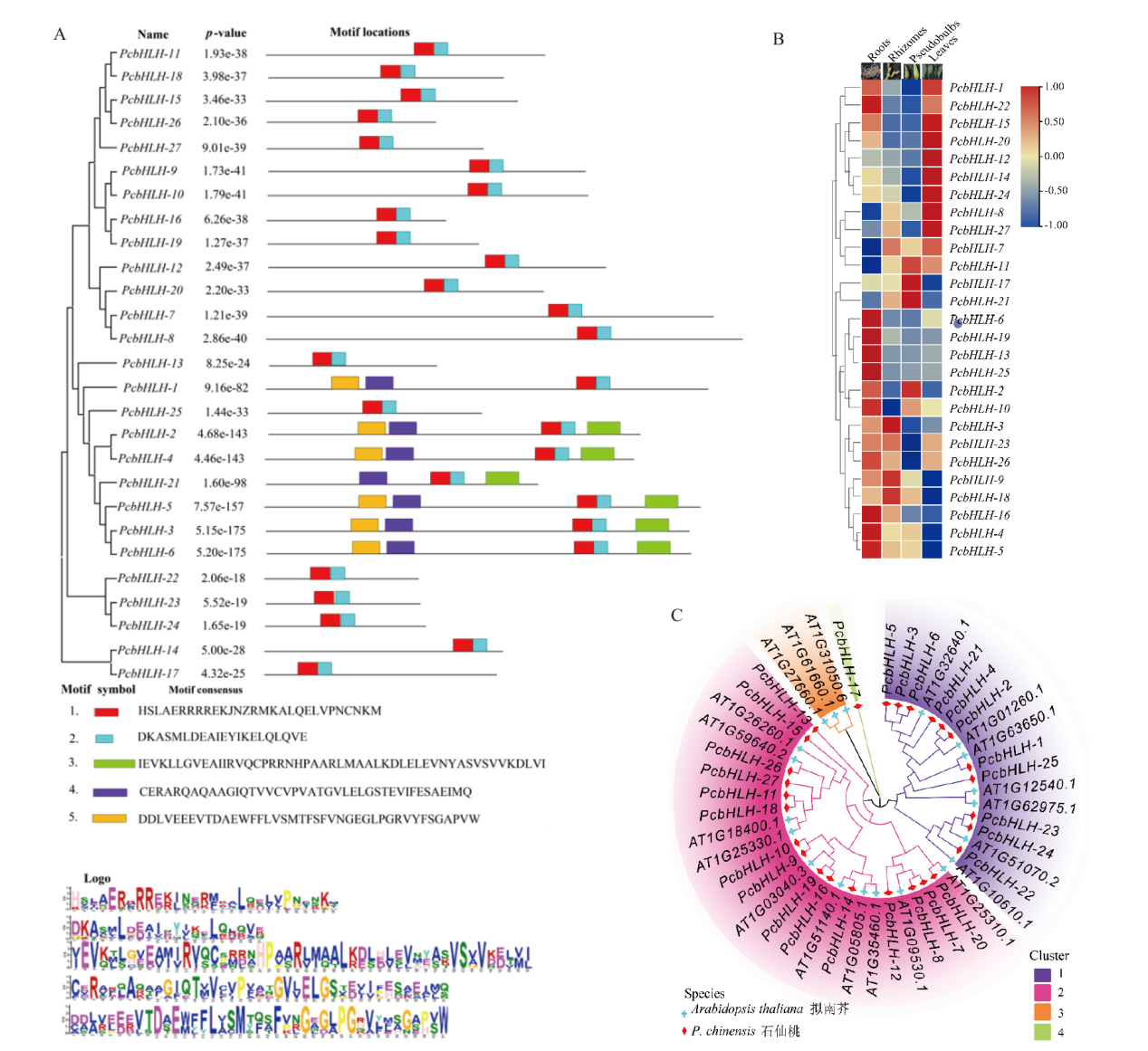
Fig. 3 Characteristics of bHLH gene family in P. chinensis A: Conserved motifs in bHLH family. B: Differential expressions of bHLH family in different tissues. C: bHLH family evolution

Fig. 4 Characteristics of WRKY gene family in P. chinensis A: Conserved motifs of WRKY family. B: WRKY family evolution. C: Differential expressions of WRKY family in different tissues

Fig. 5 Characteristics of bZIP gene family in P. chinensis A: Conserved motifs of bZIP family; B: bZIP family evolution; C: differential expressions of bZIP family in different tissues
| [1] | Liu BC, Chen JY, Zhang WJ, et al. The gastrodin biosynthetic pathway in Pholidota chinensis Lindl. revealed by transcriptome and metabolome profiling[J]. Front Plant Sci, 2022, 13: 1024239. |
| [2] | 刘保财, 黄颖桢, 赵云青, 等. 野生石仙桃立地环境与生长特性研究[J]. 福建农业学报, 2017, 32(8): 864-869. |
| Liu BC, Huang YZ, Zhao YQ, et al. Natural habitat and growth characteristics of wild Pholidota chinensis lindl[J]. Fujian J Agric Sci, 2017, 32(8): 864-869. | |
| [3] | Wang DL, Lin FX, Tang MM, et al. Two new phenanthrene derivatives from Pholidota chinensis[J]. Phytochem Lett, 2021, 43: 123-125. |
| [4] | Li DL, Zheng XL, Duan L, et al. Ethnobotanical survey of herbal tea plants from the traditional markets in Chaoshan, China[J]. J Ethnopharmacol, 2017, 205: 195-206. |
| [5] | Ti HH, Zhuang ZX, Li Y, et al. Three new phenanthrenes from Pholidota chinensis Lindl. and their antibacterial activity[J]. Nat Prod Res, 2022, 36(8): 2056-2062. |
| [6] | 袁佳, 张苑雯, 遆慧慧, 等. 石仙桃化学成分及其抗菌抗炎活性研究[J]. 广东药科大学学报, 2021, 37(4): 1-7. |
| Yuan J, Zhang YW, Ti HH, et al. Study on the chemical constituents, antimicrobial and anti-inflammatory activities of Pholidota chinensis Lindl[J]. J Guangdong Pharm Univ, 2021, 37(4): 1-7. | |
| [7] | Luo DH, Wang ZJ, Li ZM, et al. Structure of an entangled heteropolysaccharide from Pholidota chinensis Lindl and its antioxidant and anti-cancer properties[J]. Int J Biol Macromol, 2018, 112: 921-928. |
| [8] |
Xu M, Wang AD. Recent advances in the regulation of climacteric fruit ripening: hormone, transcription factor and epigenetic modifications[J]. Front Agric Sci Eng, 2021, 8(2): 314-334.
doi: 10.15302/J-FASE-2021386 |
| [9] | Hong JC. General aspects of plant transcription factor families[M]// Plant Transcription Factors. Amsterdam: Elsevier, 2016: 35-56. |
| [10] | Wang C, Hao XL, Wang Y, et al. Identification of WRKY transcription factors involved in regulating the biosynthesis of the anti-cancer drug camptothecin in Ophiorrhiza pumila[J]. Hortic Res, 2022, 9: uhac099. |
| [11] | Yang LY, Miao LY, Gong Q, et al. Advances in studies on transcription factors in regulation of secondary metabolites in Chinese medicinal plants[J]. Plant Cell Tissue Organ Cult PCTOC, 2022, 151(1): 1-9. |
| [12] |
刘保财, 胡学博, 张武君, 等. 石仙桃与细叶石仙桃叶绿体基因组解析及其系统发育[J]. 草业学报, 2024, 33(4): 171-185.
doi: 10.11686/cyxb2023192 |
| Liu BC, Hu XB, Zhang WJ, et al. Chloroplast genome assembly and phylogenetic analysis of Pholidota chinensis and Pholidota cantonensis[J]. Acta Prataculturae Sinica, 2024, 33(4): 171-185. | |
| [13] |
Zheng Y, Jiao C, Sun HH, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases[J]. Mol Plant, 2016, 9(12): 1667-1670.
doi: S1674-2052(16)30223-4 pmid: 27717919 |
| [14] | 刘美琦, 孙伟, 孟祥霄, 等. 药用植物大麻C2H2基因家族鉴定与表达分析[J]. 药学学报, 2021, 56(5): 1486-1496. |
| Liu MQ, Sun W, Meng XX, et al. Identification and expression analysis of the C2H2 gene family in Cannabis sativa L[J]. Acta Pharm Sin, 2021, 56(5): 1486-1496. | |
| [15] |
陈凤琼, 陈秋森, 林佳昕, 等. 番茄DIR基因家族鉴定及其对非生物胁迫响应的分析[J]. 中国农业科学, 2022, 55(19): 3807-3824.
doi: 10.3864/j.issn.0578-1752.2022.19.010 |
| Chen FQ, Chen QS, Lin JX, et al. Genome-wide identification of DIR family genes in tomato and response to abiotic stress[J]. Sci Agric Sin, 2022, 55(19): 3807-3824. | |
| [16] |
赵玎玲, 王梦璇, 孙天杰, 等. 大豆单锌指蛋白基因GmSZFP的克隆及其在SMV与寄主互作中的功能[J]. 中国农业科学, 2022, 55(14): 2685-2695.
doi: 10.3864/j.issn.0578-1752.2022.14.001 |
| Zhao DL, Wang MX, Sun TJ, et al. Cloning of the soybean single zinc finger protein gene GmSZFP and its functional analysis in SMV-host interactions[J]. Sci Agric Sin, 2022, 55(14): 2685-2695. | |
| [17] | 赵霞, 李元敏, 李依民, 等. 基于全长转录组测序的掌叶大黄R1-MYB基因家族鉴定分析[J]. 药学学报, 2023, 58(5): 1354-1363. |
| Zhao X, Li YM, Li YM, et al. Identification and analysis of R1-MYB gene family in Rheum palmatum L. based on full-length transcriptome sequencing[J]. Acta Pharm Sin, 2023, 58(5): 1354-1363. | |
| [18] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [19] | 钟婵娟, 彭伟业, 王冰, 等. 植物逆境响应相关的C2H2型锌指蛋白研究进展[J]. 植物生理学报, 2020, 56(11): 2356-2366. |
| Zhong CJ, Peng WY, Wang B, et al. Advances of plant C2H2 zinc finger proteins in response to stresses[J]. Plant Physiol J, 2020, 56(11): 2356-2366. | |
| [20] | Bai QX, Niu ZM, Chen QY, et al. The C2H2-type zinc finger transcription factor OSIC1 positively regulates stomatal closure under osmotic stress in poplar[J]. Plant Biotechnol J, 2023, 21(5): 943-960. |
| [21] | Feng Y, Zhang SR, Li J, et al. Dual-function C2H2-type zinc-finger transcription factor GmZFP7 contributes to isoflavone accumulation in soybean[J]. New Phytol, 2023, 237(5): 1794-1809. |
| [22] | Mohammad, Hurrah IM, Kumar A, et al. Synergistic interaction of two bHLH transcription factors positively regulates artemisinin biosynthetic pathway in Artemisia annua L[J]. Physiol Plant, 2023, 175(1): e13849. |
| [23] | Wang WY, Yu JQ, Du MC, et al. Basic helix-loop-helix(bHLH)transcription factor MdbHLH3 negatively affects the storage performance of postharvest apple fruit[J]. Hortic Plant J, 2022, 8(6): 700-712. |
| [24] | 田韦韦, 严志祥, 王成, 等. 基于全长转录组测序的多花黄精WRKY转录因子家族分析[J]. 中国中药杂志, 2023, 48(4): 939-950. |
| Tian WW, Yan ZX, Wang C, et al. Analysis of WRKY transcription factor family based on full-length transcriptome sequencing in Polygonatum cyrtonema[J]. China J Chin Mater Med, 2023, 48(4): 939-950. | |
| [25] | 叶炜, 王培育, 颜沛沛, 等. 文心兰WRKY转录因子在印度梨形孢诱导的抗软腐病作用中的响应分析[J]. 植物生理学报, 2023, 59(1): 113-126. |
| Ye W, Wang PY, Yan PP, et al. Response analysis of WRKY transcription factors in resistance to soft rot induced by Piriformospora indica in Oncidium[J]. Plant Physiol J, 2023, 59(1): 113-126. | |
| [26] | Ren LP, Wan WY, Yin DD, et al. Genome-wide analysis of WRKY transcription factor genes in Toona sinensis: an insight into evolutionary characteristics and terpene synthesis[J]. Front Plant Sci, 2023, 13: 1063850. |
| [27] |
崔荣秀, 张议文, 陈晓倩, 等. 植物bZIP参与胁迫应答调控的最新研究进展[J]. 生物技术通报, 2019, 35(2): 143-155.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0562 |
| Cui RX, Zhang YW, Chen XQ, et al. The latest research progress on the stress responses of bZIP involved in plants[J]. Biotechnol Bull, 2019, 35(2): 143-155. | |
| [28] | Niu SK, Gu XY, Zhang Q, et al. Grapevine bZIP transcription factor bZIP45 regulates VvANN1 and confers drought tolerance in Arabidopsis[J]. Front Plant Sci, 2023, 14: 1128002. |
| [29] | 刘保财, 陈菁瑛, 黄颖桢, 等. 石仙桃人工栽培技术[J]. 福建农业科技, 2017(12): 40-42. |
| Liu BC, Chen JY, Huang YZ, et al. Artificial cultivation techniques of Pholidota chinensis Lindl[J]. Fujian Agric Sci Technol, 2017(12): 40-42. | |
| [30] |
张桐, 李智强, 伍国强. WRKY转录因子在植物逆境响应中的作用[J]. 生物技术通报, 2021, 37(10): 203-215.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1481 |
|
Zhang T, Li ZQ, Wu GQ. Role of WRKY transcription factor in plant response to stresses[J]. Biotechnol Bull, 2021, 37(10): 203-215.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1481 |
|
| [31] |
Kumar R, Das S, Mishra M, et al. Emerging roles of NAC transcription factor in medicinal plants: progress and prospects[J]. 3 Biotech, 2021, 11(10): 425.
doi: 10.1007/s13205-021-02970-x pmid: 34567930 |
| [32] | Li C, Gong QQ, Liu P, et al. Co-expressed network analysis based on 289 transcriptome samples reveals methyl jasmonate-mediated gene regulatory mechanism of flavonoid compounds in Dendrobium catenatum[J]. Plant Physiol Biochem, 2024, 206: 108226. |
| [33] |
Zeng X, Ling H, Chen XM, et al. Genome-wide identification, phylogeny and function analysis of GRAS gene family in Dendrobium catenatum(Orchidaceae)[J]. Gene, 2019, 705: 5-15.
doi: S0378-1119(19)30391-9 pmid: 30999026 |
| [34] | Wang MJ, Ou Y, Li Z, et al. Genome-wide identification and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Cymbidium ensifolium[J]. Int J Mol Sci, 2023, 24(4): 3825. |
| [35] | Yang K, Hou YB, Wu M, et al. DoMYB5 and DobHLH24, transcription factors involved in regulating anthocyanin accumulation in Dendrobium officinale[J]. Int J Mol Sci, 2023, 24(8): 7552. |
| [36] | Chuang YC, Hung YC, Tsai WC, et al. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids[J]. J Exp Bot, 2018, 69(18): 4363-4377. |
| [37] | Hsieh MH, Pan ZJ, Lai PH, et al. Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids[J]. J Exp Bot, 2013, 64(12): 3869-3884. |
| [38] | 邢丙聪, 苏立样, 万思琦, 等. 金线莲中调控胚胎发育WRKY转录因子筛选及克隆分析[J]. 中草药, 2022, 53(12): 3745-3754. |
| Xing BC, Su LY, Wan SQ, et al. Screening and cloning analysis of WRKY transcription factor regulating embryo development from Anoectochilus roxburghii[J]. Chin Tradit Herb Drugs, 2022, 53(12): 3745-3754. | |
| [39] | Zeng DQ, Teixeira da Silva JA, Zhang MZ, et al. Genome-wide identification and analysis of the APETALA2(AP2)transcription factor in Dendrobium officinale[J]. Int J Mol Sci, 2021, 22(10): 5221. |
| [1] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [2] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [3] | LI Can, JIANG Xiang-ning, GAI Ying. Cloning of the LkF3H2 Gene in Larix kaempferi and Its Function in Regulating Flavonoid Metabolism [J]. Biotechnology Bulletin, 2024, 40(2): 245-252. |
| [4] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| [5] | WANG Bin, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui, HE Jin-ming. Cloning of bHLH96 Gene and Its Roles in Regulating the Biosynthesis of Peppermint Terpenes [J]. Biotechnology Bulletin, 2024, 40(1): 281-293. |
| [6] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [7] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [8] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [9] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [10] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [11] | CHEN Chu-wen, LI Jie, ZHAO Rui-peng, LIU Yuan, WU Jin-bo, LI Zhi-xiong. Cloning, Tissue Expression Profile and Function Prediction of GPX3 Gene in Tibetan Chicken [J]. Biotechnology Bulletin, 2023, 39(3): 311-320. |
| [12] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [13] | YAN Meng-yu, WEI Xiao-wei, CAO Jing, LAN Hai-yan. Cloning of Basic Helix-loop-helix(bHLH)Transcription Factor Gene SabHLH169 in Suaeda aralocaspica and Analysis of Its Resistances to Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 328-339. |
| [14] | AN Chang, LU Lin, SHEN Meng-qian, CHEN Sheng-zhen, YE Kang-zhuo, QIN Yuan, ZHENG Ping. Research Progress of bHLH Gene Family in Plants and Its Application Prospects in Medical Plants [J]. Biotechnology Bulletin, 2023, 39(10): 1-16. |
| [15] | CHEN Hao-ting, ZHANG Yu-jing, LIU Jie, DAI Ze-min, LIU Wei, SHI Yu, ZHANG Yi, LI Tian-lai. Functional Analysis of WRKY6 Gene in Tomato Under Low-phosphorus Stress [J]. Biotechnology Bulletin, 2023, 39(10): 136-147. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||