








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 49-61.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0506
Previous Articles Next Articles
TAN Jing-xuan( ), XING De-xun, HE Tian-jin, LIU Zhan-ying(
), XING De-xun, HE Tian-jin, LIU Zhan-ying( )
)
Received:2024-05-29
Online:2025-01-26
Published:2025-01-22
Contact:
LIU Zhan-ying
E-mail:tjx13345135159@163.com;hgxylzy2008@imut.edu.cn
TAN Jing-xuan, XING De-xun, HE Tian-jin, LIU Zhan-ying. Advances in Protein Expression System of Pseudomonas fluorescens[J]. Biotechnology Bulletin, 2025, 41(1): 49-61.
| 表达载体 Expression vector | 复制子 Replicon | 启动子 Promoter | 蓝白斑筛选 Blue-white screening | 诱导物 Inducer | 抗性标记 Resistance marker | 特征 Characteristics | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| pCN51 | pPS10 | Ptac, Plac | No | IPTG by Plac | Kana | 适用于多种假单胞菌 | [ |
| pBBR1MCS | pBBR1 | Ptac, Plac | Yes | IPTG by Plac | Cm | 适用多种启动子系统 | [ |
| pEX18Gm | pEX18 | lacZ | Yes | IPTG | Gm | 基因敲除和替换载体 | [ |
| pQF50 | ColE1 | lacZ | Yes | IPTG | Amp | 基于lac启动子系统 | [ |
| pET series | ColE1 | PT7 | No | IPTG | Amp | 需配合T7 RNA聚合酶 | [ |
| pEB8 | RSF1010 | PT7 | No | IPTG | Amp | 适用于含T7 RNA聚合酶的宿主菌株 | [ |
| pPLGN1 | RSF1010 | λPR | No | Temperature | Kana | 温度调控表达 | [ |
| pLV vectors | RSF1010 | λPL | No | Temperature | Kana | 温度调控表达 | [ |
| pERD20/pERD21 | RSF1010 | Pm | No | Benzoate | Kana | 苯甲酸诱导 | [ |
| pVLT vectors | RSF1010 | Ptac | No | IPTG | Kana | IPTG诱导表达 | [ |
| pMMB66EH | RSF1010 | Ptac | No | IPTG | Amp | IPTG诱导表达 | [ |
| pJB653 and derivatives | RK2 | Pm | No | m-Toluic acid | Kana | m-甲苯酸诱导 | [ |
| pRK415 | RK2 | Plac | Yes | IPTG | Tet | 具有蓝白筛选功能 | [ |
| pUCP vectors | pRO1600 | Plac | Yes | IPTG | Carb | 具有蓝白筛选功能 | [ |
| pUCPKS/pUCPSK | pRO1600 | PT7, Plac | Yes | IPTG | Carb | 双启动子系统 | [ |
| pBSP II KS/pBSP II KS | pRO1600 | PT7, Plac | Yes | IPTG | Amp | 双启动子系统 | [ |
Table 1 Common expression vectors for P. fluorescens
| 表达载体 Expression vector | 复制子 Replicon | 启动子 Promoter | 蓝白斑筛选 Blue-white screening | 诱导物 Inducer | 抗性标记 Resistance marker | 特征 Characteristics | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| pCN51 | pPS10 | Ptac, Plac | No | IPTG by Plac | Kana | 适用于多种假单胞菌 | [ |
| pBBR1MCS | pBBR1 | Ptac, Plac | Yes | IPTG by Plac | Cm | 适用多种启动子系统 | [ |
| pEX18Gm | pEX18 | lacZ | Yes | IPTG | Gm | 基因敲除和替换载体 | [ |
| pQF50 | ColE1 | lacZ | Yes | IPTG | Amp | 基于lac启动子系统 | [ |
| pET series | ColE1 | PT7 | No | IPTG | Amp | 需配合T7 RNA聚合酶 | [ |
| pEB8 | RSF1010 | PT7 | No | IPTG | Amp | 适用于含T7 RNA聚合酶的宿主菌株 | [ |
| pPLGN1 | RSF1010 | λPR | No | Temperature | Kana | 温度调控表达 | [ |
| pLV vectors | RSF1010 | λPL | No | Temperature | Kana | 温度调控表达 | [ |
| pERD20/pERD21 | RSF1010 | Pm | No | Benzoate | Kana | 苯甲酸诱导 | [ |
| pVLT vectors | RSF1010 | Ptac | No | IPTG | Kana | IPTG诱导表达 | [ |
| pMMB66EH | RSF1010 | Ptac | No | IPTG | Amp | IPTG诱导表达 | [ |
| pJB653 and derivatives | RK2 | Pm | No | m-Toluic acid | Kana | m-甲苯酸诱导 | [ |
| pRK415 | RK2 | Plac | Yes | IPTG | Tet | 具有蓝白筛选功能 | [ |
| pUCP vectors | pRO1600 | Plac | Yes | IPTG | Carb | 具有蓝白筛选功能 | [ |
| pUCPKS/pUCPSK | pRO1600 | PT7, Plac | Yes | IPTG | Carb | 双启动子系统 | [ |
| pBSP II KS/pBSP II KS | pRO1600 | PT7, Plac | Yes | IPTG | Amp | 双启动子系统 | [ |
| 信号肽 Signal peptide | 来源 Origin | 功能 Function | 应用实例 Application | 表达特性 Characteristics | 参考文献 Reference |
|---|---|---|---|---|---|
| PelB | 枯草芽胞杆菌的果胶酶B | 引导重组蛋白通过Sec途径分泌至周质空间 | G-CSF(人粒细胞集落刺激因子) | 可溶,活性,96孔板规模为250 mg/L | [ |
| OmpA | 大肠杆菌的外膜蛋白A | 引导外膜蛋白的分泌 | G-CSF(人粒细胞集落刺激因子) | 可溶,活性,96孔板规模为250 mg/L | [ |
| PhoA | 大肠杆菌的碱性磷酸酶 | 引导碱性磷酸酶的分泌 | Fab | 可溶,活性,100 mg/L | [ |
| TorA | 大肠杆菌的甲基氨基甲酸酯还原酶 | 专用于Tat途径,适合复杂酶类的分泌 | 人血清白蛋白(HSA) | 可溶,活性,1.5 g/L | [ |
| LipA | 荧光假单胞菌的脂肪酶 | 引导脂肪酶的分泌 | 重组脂肪酶(recombinant lipase) | 可溶,活性,100 mg/L | [ |
| AmyL | 枯草芽胞杆菌的α-淀粉酶 | 引导α-淀粉酶的分泌 | 重组α-淀粉酶 | 可溶,活性,85 g/L | [ |
| DsbA | 大肠杆菌的二硫键异构酶 | 本身是一种二硫键异构酶,帮助新合成蛋白形成正确的二硫键 | 人类生长激素(hGH) | 可溶,活性,1.2 g/L | [ |
| HlyA | 大肠杆菌的血红素结合蛋白 | 通过I型分泌系统引导分泌血红素结合蛋白 | 血红素结合蛋白 | 可溶,活性,产量未知 | [ |
| AprA | 荧光假单胞菌的碱性蛋白酶 | 引导碱性蛋白酶的分泌 | 碱性蛋白酶 | 可溶,活性,产量未知 | [ |
Table 2 Application of signal peptides in P. fluorescens
| 信号肽 Signal peptide | 来源 Origin | 功能 Function | 应用实例 Application | 表达特性 Characteristics | 参考文献 Reference |
|---|---|---|---|---|---|
| PelB | 枯草芽胞杆菌的果胶酶B | 引导重组蛋白通过Sec途径分泌至周质空间 | G-CSF(人粒细胞集落刺激因子) | 可溶,活性,96孔板规模为250 mg/L | [ |
| OmpA | 大肠杆菌的外膜蛋白A | 引导外膜蛋白的分泌 | G-CSF(人粒细胞集落刺激因子) | 可溶,活性,96孔板规模为250 mg/L | [ |
| PhoA | 大肠杆菌的碱性磷酸酶 | 引导碱性磷酸酶的分泌 | Fab | 可溶,活性,100 mg/L | [ |
| TorA | 大肠杆菌的甲基氨基甲酸酯还原酶 | 专用于Tat途径,适合复杂酶类的分泌 | 人血清白蛋白(HSA) | 可溶,活性,1.5 g/L | [ |
| LipA | 荧光假单胞菌的脂肪酶 | 引导脂肪酶的分泌 | 重组脂肪酶(recombinant lipase) | 可溶,活性,100 mg/L | [ |
| AmyL | 枯草芽胞杆菌的α-淀粉酶 | 引导α-淀粉酶的分泌 | 重组α-淀粉酶 | 可溶,活性,85 g/L | [ |
| DsbA | 大肠杆菌的二硫键异构酶 | 本身是一种二硫键异构酶,帮助新合成蛋白形成正确的二硫键 | 人类生长激素(hGH) | 可溶,活性,1.2 g/L | [ |
| HlyA | 大肠杆菌的血红素结合蛋白 | 通过I型分泌系统引导分泌血红素结合蛋白 | 血红素结合蛋白 | 可溶,活性,产量未知 | [ |
| AprA | 荧光假单胞菌的碱性蛋白酶 | 引导碱性蛋白酶的分泌 | 碱性蛋白酶 | 可溶,活性,产量未知 | [ |
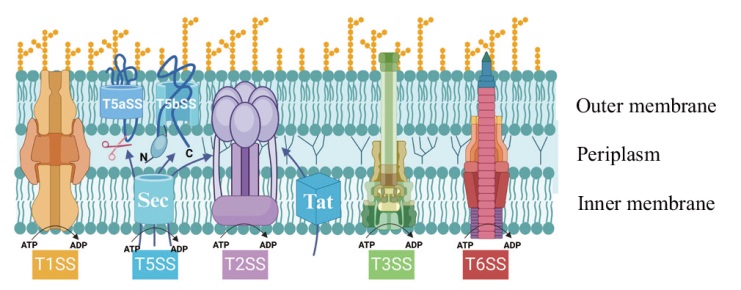
Fig. 1 Schematic representation of the different secretion systems in P. fluorescens T1SS: Type I secretion system. T2SS: Type II secretion system. T3SS: Type III secretion system. T5SS: Type V secretion system. T5aSS: Autotransporter system. T5bSS: Two-partner secretion system. Sec: Sec secretion system. Tat: Tat secretion system. T6SS: Type VI secretion system
| 分泌系统 Secretion system | 膜锚定位置Membrane anchoring site | 存在种属Species present | 参考文献 Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IM | OM | G- | G+ | Arch | Euk | ||||
| Sec secretory pathway | + | - | + | + | + | + | [ | ||
| Twin arginine targeting(Tat)pathway | + | - | + | + | + | + | [ | ||
| Holins | + | - | + | + | + | - | [ | ||
| Large conductance mechanosensitive ion channel(MscL) | + | - | + | + | + | + | [ | ||
| Outer membrane FUP | - | + | + | + | - | - | [ | ||
| Type I secretion system(ABC) | + | - | + | + | - | + | [ | ||
| Type II secretion system(MTB) | + | - | + | + | - | - | [ | ||
| Type III secretion system | + | - | + | - | - | - | [ | ||
| Type V secretion system(AT) | + | - | + | - | - | - | [ | ||
| Type VI secretion system | + | - | + | - | - | - | [ | ||
Table 3 Characterized secretion systems and secreted proteins in P. fluorescens
| 分泌系统 Secretion system | 膜锚定位置Membrane anchoring site | 存在种属Species present | 参考文献 Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IM | OM | G- | G+ | Arch | Euk | ||||
| Sec secretory pathway | + | - | + | + | + | + | [ | ||
| Twin arginine targeting(Tat)pathway | + | - | + | + | + | + | [ | ||
| Holins | + | - | + | + | + | - | [ | ||
| Large conductance mechanosensitive ion channel(MscL) | + | - | + | + | + | + | [ | ||
| Outer membrane FUP | - | + | + | + | - | - | [ | ||
| Type I secretion system(ABC) | + | - | + | + | - | + | [ | ||
| Type II secretion system(MTB) | + | - | + | + | - | - | [ | ||
| Type III secretion system | + | - | + | - | - | - | [ | ||
| Type V secretion system(AT) | + | - | + | - | - | - | [ | ||
| Type VI secretion system | + | - | + | - | - | - | [ | ||
| [1] | Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond[J]. Biotechnol Adv, 2012, 30(5): 1102-1107. |
| [2] | Pang J, Wang JS, Liu ZY, et al. Identification and characterization of an Endo-glucanase secreted from cellulolytic Escherichia coli ZH-4[J]. BMC Biotechnol, 2019, 19(1): 63. |
| [3] | Pang J, Liu ZY, Zhang QC, et al. Systematic analysis of Escherichia coli isolates from sheep and cattle suggests adaption to the rumen niche[J]. Appl Environ Microbiol, 2020, 86(20): e01417-20. |
| [4] | Retallack DM, Jin HF, Chew L. Reliable protein production in a Pseudomonas fluorescens expression system[J]. Protein Expr Purif, 2012, 81(2): 157-165. |
| [5] | Ma QH, Zhai YF, Schneider JC, et al. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens[J]. Biochim Biophys Acta Biomembr, 2003, 1611(1/2): 223-233. |
| [6] | Gupta G, Chauhan PS, Jha PN, et al. Secretory molecules from secretion systems fine-tune the host-beneficial bacteria(PGPRs)interaction[J]. Front Microbiol, 2024, 15: 1355750. |
| [7] | Gallique M, Decoin V, Barbey C, et al. Contribution of the Pseu-domonas fluorescens MFE01 type VI secretion system to biofilm formation[J]. PLoS One, 2017, 12(1): e0170770. |
| [8] | Squires CH, Retallack DM, Chew LC, et al. Heterologous protein production in P. fluorescens[J]. BioProcess Int, 2004, 2: 54-59. |
| [9] | Son M, Moon Y, Oh MJ, et al. Lipase and protease double-deletion mutant of Pseudomonas fluorescens suitable for extracellular protein production[J]. Appl Environ Microbiol, 2012, 78(23): 8454-8462. |
| [10] | Schneider JC, Jenings AF, Mun DM, et al. Auxotrophic markers pyrF and proC can replace antibiotic markers on protein production plasmids in high-cell-density Pseudomonas fluorescens fermentation[J]. Biotechnol Prog, 2005, 21(2): 343-348. |
| [11] | Fabia BU, Bingwa J, Park J, et al. Utilizing the abc transporter for growth factor production by fleQ deletion mutant of Pseudomonas fluorescens[J]. Biomedicines, 2021, 9(6): 679. |
| [12] | Abolmaaty A, Abdelkader RMM, Amin DH. Synergistic inhibition of Pseudomonas fluorescens growth and proteases activities via sodium chlorite-based oxyhalogen[J]. World J Microbiol Biotechnol, 2022, 39(1): 33. |
| [13] | Byun H, Park J, Fabia BU, et al. Generalized approach towards secretion-based protein production via neutralization of secretion-preventing cationic substrate residues[J]. Int J Mol Sci, 2022, 23(12): 6700. |
| [14] | Dodge AG, Thoma CJ, O'Connor MR, et al. Recombinant Pseu-domonas growing on non-natural fluorinated substrates shows stress but overall tolerance to cytoplasmically released fluoride anion[J]. mBio, 2024, 15(1): e0278523. |
| [15] | Oku S, Komatsu A, Tajima T, et al. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization[J]. Microbes Environ, 2012, 27(4): 462-469. |
| [16] | Decoin V, Gallique M, Barbey C, et al. A Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility and bacterial competition[J]. BMC Microbiol, 2015, 15: 72. |
| [17] | Huong DDT, Rajalingam N, Lee YH. Characterization of virulence function of Pseudomonas cichorii avirulence protein E1(AvrE1)during host plant infection[J]. Plant Pathol J, 2021, 37(5): 494-501. |
| [18] | Trögl J, Chauhan A, Ripp S, et al. Pseudomonas fluorescens HK44: lessons learned from a model whole-cell bioreporter with a broad application history[J]. Sensors(Basel), 2012, 12(2): 1544-1571. |
| [19] | Pastora AB, Rzasa KM, O'Toole GA. Multiple pathways impact the swarming motility of Pseudomonas fluorescens Pf0-1[J]. Microbiol Spectr, 2024, 12(6): e00166-24. |
| [20] | Kim W, Silby MW, Purvine SO, et al. Proteomic detection of non-annotated protein-coding genes in Pseudomonas fluorescens Pf0-1[J]. PLoS One, 2009, 4(12): e8455. |
| [21] | Silby MW, Cerdeño-Tárraga AM, Vernikos GS, et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens[J]. Genome Biol 2009, 10(5): R51. |
| [22] | Koza A, Kusmierska A, McLaughlin K, et al. Adaptive radiation of Pseudomonas fluorescens SBW25 in experimental microcosms provides an understanding of the evolutionary ecology and molecular biology of A-L interface biofilm formation[J]. FEMS Microbiol Lett, 2017, 364(12). DOI: 10.1093/femsle/fnx109. |
| [23] |
Theodosiou L, Farr AD, Rainey PB. Barcoding populations of Pseu-domonas fluorescens SBW25[J]. J Mol Evol, 2023, 91(3): 254-262.
doi: 10.1007/s00239-023-10103-6 pmid: 37186220 |
| [24] | Moshynets OV, Pokholenko I, Iungin O, et al. EDNA, amyloid fibers and membrane vesicles identified in Pseudomonas fluorescens SBW25 biofilms[J]. Int J Mol Sci, 2022, 23(23): 15096. |
| [25] |
Gallie J, Bertels F, Remigi P, et al. Repeated phenotypic evolution by different genetic routes in Pseudomonas fluorescens SBW25[J]. Mol Biol Evol, 2019, 36(5): 1071-1085.
doi: 10.1093/molbev/msz040 pmid: 30835268 |
| [26] | Wu X, Monchy S, Taghavi S, et al. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida[J]. FEMS Microbiol Rev, 2011, 35(2): 299-323. |
| [27] | Lim CK, Hassan KA, Tetu SG, et al. The effect of iron limitation on the transcriptome and proteome of Pseudomonas fluorescens Pf-5[J]. PLoS One, 2012, 7(6): e39139. |
| [28] | Garrido-Sanz D, Redondo-Nieto M, Martin M, et al. Comparative genomics of the Pseudomonas corrugata subgroup reveals high species diversity and allows the description of Pseudomonas ogarae sp. nov[J]. Microb Genom, 2021, 7(6): 000593. |
| [29] | Liu XX, Xu J, Zhu JL, et al. Combined transcriptome and proteome analysis of RpoS regulon reveals its role in spoilage potential of Pseudomonas fluorescens[J]. Front Microbiol, 2019, 10: 94. |
| [30] | Nakashima N, Tamura T. Cell-free protein synthesis using cell extract of Pseudomonas fluorescens and CspA promoter[J]. Biochem Biophys Res Commun, 2004, 319(2): 671-676. |
| [31] |
Sun MM, Gao AX, Liu XX, et al. High-throughput process development from gene cloning to protein production[J]. Microb Cell Fact, 2023, 22(1): 182.
doi: 10.1186/s12934-023-02184-1 pmid: 37715258 |
| [32] |
Nieto C, Fernández-Tresguerres E, Sánchez N, et al. Cloning vectors, derived from a naturally occurring plasmid of Pseudomonas savastanoi, specifically tailored for genetic manipulations in Pseu-domonas[J]. Gene, 1990, 87(1): 145-149.
pmid: 2110095 |
| [33] |
Kovach ME, Elzer PH, Hill DS, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes[J]. Gene, 1995, 166(1): 175-176.
doi: 10.1016/0378-1119(95)00584-1 pmid: 8529885 |
| [34] | Ge YH, Pei DL, Feng PY, et al. Autoinduction of RpoS biosynthesis in the biocontrol strain Pseudomonas sp. M18[J]. Curr Microbiol, 2007, 54(2): 97-101. |
| [35] |
Farinha MA, Kropinski AM. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters[J]. J Bacteriol, 1990, 172(6): 3496-3499.
pmid: 2111810 |
| [36] |
Shilling PJ, Mirzadeh K, Cumming AJ, et al. Improved designs for pET expression plasmids increase protein production yield in Es-cherichia coli[J]. Commun Biol, 2020, 3(1): 214.
doi: 10.1038/s42003-020-0939-8 pmid: 32382055 |
| [37] | Broto A, Gaspari E, Miravet-Verde S, et al. A genetic toolkit and gene switches to limit Mycoplasma growth for biosafety applications[J]. Nat Commun, 2022, 13(1): 1910. |
| [38] | Volke DC, Friis L, Wirth NT, et al. Synthetic control of plasmid replication enables target- and self-curing of vectors and expedites genome engineering of Pseudomonas putida[J]. Metab Eng Commun, 2020, 10: e00126. |
| [39] | Weihmann R, Kubicki S, Bitzenhofer NL, et al. The modular pYT vector series employed for chromosomal gene integration and expression to produce carbazoles and glycolipids in P. putida[J]. FEMS Microbes, 2022, 4: xtac030. |
| [40] | Alker AT, Farrell MV, Aspiras AE, et al. A modular plasmid toolkit applied in marine bacteria reveals functional insights during bacteria-stimulated metamorphosis[J]. mBio, 2023, 14(4): e0150223. |
| [41] |
Wichmann J, Behrendt G, Boecker S, et al. Characterizing and utilizing oxygen-dependent promoters for efficient dynamic metabolic engineering[J]. Metab Eng, 2023, 77: 199-207.
doi: 10.1016/j.ymben.2023.04.006 pmid: 37054967 |
| [42] | Kato R, Maekawa K, Kobayashi S, et al. Stereoinversion via alcohol dehydrogenases enables complete catabolism of β-1-type lignin-derived aromatic isomers[J]. Appl Environ Microbiol, 2023, 89(6): e0017123. |
| [43] | Choi Y, Joo M, Song W, et al. Transcript-specific selective translation by specialized ribosomes bearing genome-encoded heterogeneous rRNAs in V. vulnificus CMCP6[J]. J Microbiol, 2022, 60(12): 1162-1167. |
| [44] | Kotecka K, Kawalek A, Modrzejewska-Balcerek M, et al. Functional characterization of TetR-like transcriptional regulator PA3973 from Pseudomonas aeruginosa[J]. Int J Mol Sci, 2022, 23(23): 14584. |
| [45] | Moraskie M, Roshid MHO, O'Connor G, et al. Microbial whole-cell biosensors: current applications, challenges, and future perspectives[J]. Biosens Bioelectron, 2021, 191: 113359. |
| [46] | Chen X, Kaiser CM. AP profiling resolves co-translational folding pathways and chaperone interactions in vivo[J]. Biophys J, 123(3): 301A-301A. |
| [47] | Jorth P, McLean K, Ratjen A, et al. Evolved aztreonam resistance is multifactorial and can produce hypervirulence in Pseudomonas aeruginosa[J]. mBio, 2017, 8(5): e00517-17. |
| [48] | Lucas S, Toffin L, Zivanovic Y, et al. Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi[J]. Appl Environ Microbiol, 2002, 68(11): 5528-5536. |
| [49] |
Boontawon T, Nakazawa T, Inoue C, et al. Efficient genome editing with CRISPR/Cas9 in Pleurotus ostreatus[J]. AMB Express, 2021, 11(1): 30.
doi: 10.1186/s13568-021-01193-w pmid: 33609205 |
| [50] | Shi TQ, Yang CL, Li DX, et al. Establishment of a selectable marker recycling system for iterative gene editing in Fusarium fujik-uroi[J]. Synth Syst Biotechnol, 2024, 9(1): 159-164. |
| [51] | Yu L, Xiao ML, Zhu ZH, et al. Efficient genome editing in Claviceps purpurea using a CRISPR/Cas9 ribonucleoprotein method[J]. Synth Syst Biotechnol, 2022, 7(2): 664-670. |
| [52] |
Retallack DM, Thomas TC, Shao Y, et al. Identification of anthranilate and benzoate metabolic operons of Pseudomonas fluorescens and functional characterization of their promoter regions[J]. Microb Cell Fact, 2006, 5: 1.
pmid: 16396686 |
| [53] |
de Pinho Favaro MT, Atienza-Garriga J, Martínez-Torró C, et al. Recombinant vaccines in 2022: a perspective from the cell factory[J]. Microb Cell Fact, 2022, 21(1): 203.
doi: 10.1186/s12934-022-01929-8 pmid: 36199085 |
| [54] | Lycklama A Nijeholt JA, Driessen AJM. The bacterial sec-translocase: structure and mechanism[J]. Philos Trans R Soc Lond B Biol Sci, 2012, 367(1592): 1016-1028. |
| [55] |
García de Viedma D, Giraldo R, Ruiz-Echevarría MJ, et al. Transcription of repA, the gene of the initiation protein of the Pseu-domonas plasmid pPS10, is autoregulated by interactions of the RepA protein at a symmetrical operator[J]. J Mol Biol, 1995, 247(2): 211-223.
pmid: 7707370 |
| [56] | Li J, Wang BY, Yang Q, et al. Enabling efficient genetic manipulations in a rare actinomycete Pseudonocardia alni Shahu[J]. Front Microbiol, 2022, 13: 848964. |
| [57] |
Yan Q, Fong SS. Challenges and advances for genetic engineering of non-model bacteria and uses in consolidated bioprocessing[J]. Front Microbiol, 2017, 8: 2060.
doi: 10.3389/fmicb.2017.02060 pmid: 29123506 |
| [58] | Kamensek U, Rencelj A, Jesenko T, et al. Maintenance and gene electrotransfer efficiency of antibiotic resistance gene-free plasmids encoding mouse, canine and human interleukin-12 orthologues[J]. Heliyon, 2022, 8(2): e08879. |
| [59] | Jin HF, Cantin GT, Maki S, et al. Soluble periplasmic production of human granulocyte colony-stimulating factor(G-CSF)in Pseu-domonas fluorescens[J]. Protein Expr Purif, 2011, 78(1): 69-77. |
| [60] | Gundinger T, Kittler S, Kubicek S, et al. Recombinant protein production in E. coli using the phoA expression system[J]. Fermentation, 2022, 8(4): 181. |
| [61] |
Retallack DM, Schneider JC, Mitchell J, et al. Transport of heterologous proteins to the periplasmic space of Pseudomonas fluorescens using a variety of native signal sequences[J]. Biotechnol Lett, 2007, 29(10): 1483-1491.
pmid: 17541504 |
| [62] | Liu XH, Viswanadhapalli S, Kumar S, et al. Targeting LIPA independent of its lipase activity is a therapeutic strategy in solid tumors via induction of endoplasmic reticulum stress[J]. Nat Cancer, 2022, 3(7): 866-884. |
| [63] |
Gupta SK, Shukla P. Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications[J]. Crit Rev Biotechnol, 2016, 36(6): 1089-1098.
pmid: 26384140 |
| [64] | Schulz E, Schumann M, Schneemann M, et al. Escherichia coli alpha-hemolysin HlyA induces host cell polarity changes, epithelial barrier dysfunction and cell detachment in human colon carcinoma caco-2 cell model via PTEN-dependent dysregulation of cell junctions[J]. Toxins, 2021, 13(8): 520. |
| [65] |
Frisoni GB, Festari C, Massa F, et al. European intersocietal recommendations for the biomarker-based diagnosis of neurocognitive disorders[J]. Lancet Neurol, 2024, 23(3): 302-312.
doi: 10.1016/S1474-4422(23)00447-7 pmid: 38365381 |
| [66] |
Palmer T, Berks BC. The twin-arginine translocation(Tat)protein export pathway[J]. Nat Rev Microbiol, 2012, 10(7): 483-496.
doi: 10.1038/nrmicro2814 pmid: 22683878 |
| [67] |
Pena RT, Blasco L, Ambroa A, et al. Relationship between quorum sensing and secretion systems[J]. Front Microbiol, 2019, 10: 1100.
doi: 10.3389/fmicb.2019.01100 pmid: 31231316 |
| [68] |
Rapisarda C, Fronzes R. Secretion systems used by bacteria to subvert host functions[J]. Curr Issues Mol Biol, 2018, 25: 1-42.
doi: 10.21775/cimb.025.001 pmid: 28875938 |
| [69] |
Costa TRD, Felisberto-Rodrigues C, Meir A, et al. Secretion systems in gram-negative bacteria: structural and mechanistic insights[J]. Nat Rev Microbiol, 2015, 13(6): 343-359.
doi: 10.1038/nrmicro3456 pmid: 25978706 |
| [70] | Durán D, Bernal P, Vazquez-Arias D, et al. Pseudomonas fluo-rescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome[J]. Sci Rep, 2021, 11(1): 5772. |
| [71] | Berks BC, Sargent F, De Leeuw E, et al. A novel protein transport system involved in the biogenesis of bacterial electron transfer chains[J]. Biochim Biophys Acta, 2000, 1459(2/3): 325-330. |
| [72] | Callaghan JD, Stella NA, Lehner KM, et al. Xylose-inducible promoter tools for Pseudomonas species and their use in implicating a role for the type II secretion system protein XcpQ in the inhibition of corneal epithelial wound closure[J]. Appl Environ Microbiol, 2020, 86(14): e00250-20. |
| [73] | Duong F, Bonnet E, Géli V, et al. The AprX protein of Pseu-domonas aeruginosa: a new substrate for the apr type I secretion system[J]. Gene, 2001, 262(1/2): 147-153. |
| [74] | Hay ID, Belousoff MJ, Lithgow T. Structural basis of type 2 secretion system engagement between the inner and outer bacterial membranes[J]. mBio, 2017, 8(5): e01344-17. |
| [75] | Qin SG, Xiao W, Zhou CM, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics[J]. Signal Transduct Target Ther, 2022, 7(1): 199. |
| [76] |
Anlauf MT, Bilsing FL, Reiners J, et al. Type 1 secretion necessitates a tight interplay between all domains of the ABC transporter[J]. Sci Rep, 2024, 14(1): 8994.
doi: 10.1038/s41598-024-59759-0 pmid: 38637678 |
| [77] | Busch A, Waksman G. Chaperone-usher pathways: diversity and pilus assembly mechanism[J]. Philos Trans R Soc Lond B Biol Sci, 2012, 367(1592): 1112-1122. |
| [78] | Senf F, Tommassen J, Koster M. Polar secretion of proteins via the Xcp type II secretion system in Pseudomonas aeruginosa[J]. Microbiology(Reading), 2008, 154(Pt 10): 3025-3032. |
| [79] |
Sethupathy S, Prasath KG, Ananthi S, et al. Proteomic analysis reveals modulation of iron homeostasis and oxidative stress response in Pseudomonas aeruginosa PAO1 by curcumin inhibiting quorum sensing regulated virulence factors and biofilm production[J]. J Proteomics, 2016, 145: 112-126.
doi: S1874-3919(16)30141-5 pmid: 27108548 |
| [80] |
Preston GM, Bertrand N, Rainey PB. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25[J]. Mol Microbiol, 2001, 41(5): 999-1014.
pmid: 11555282 |
| [81] |
Stringlis IA, Zamioudis C, Berendsen RL, et al. Type iii secretion system of beneficial rhizobacteria Pseudomonas simiae WCS417 and Pseudomonas defensor WCS374[J]. Front Microbiol, 2019, 10: 1631.
doi: 10.3389/fmicb.2019.01631 pmid: 31379783 |
| [82] | Liu P, Zhang W, Zhang LQ, et al. Supramolecular structure and functional analysis of the type III secretion system in Pseudomonas fluorescens 2P24[J]. Front Plant Sci, 2016, 6: 1190. |
| [83] | Viollet A, Pivato B, Mougel C, et al. Pseudomonas fluorescens C7R12 type III secretion system impacts mycorrhization of Medi-cago truncatula and associated microbial communities[J]. Mycorrhiza, 2017, 27(1): 23-33. |
| [84] | Marzorati F, Rossi R, Bernardo L, et al. Arabidopsis thaliana early foliar proteome response to root exposure to the rhizobacterium Pseudomonas simiae WCS417[J]. Mol Plant Microbe Interact, 2023, 36(11): 737-748. |
| [85] | Quibod IL, Grande G, Oreiro EG, et al. Rice-infecting Pseudomonas genomes are highly accessorized and harbor multiple putative virulence mechanisms to cause sheath brown rot[J]. PLoS One, 2015, 10(9): e0139256. |
| [86] |
Dalbey RE, Kuhn A. Protein Traffic in Gram-negative bacteria-how exported and secreted proteins find their way[J]. FEMS Microbiol Rev, 2012, 36(6): 1023-1045.
doi: 10.1111/j.1574-6976.2012.00327.x pmid: 22250915 |
| [87] |
Mavrodi DV, Joe A, Mavrodi OV, et al. Structural and functional analysis of the type III secretion system from Pseudomonas fluo-rescens Q8r1-96[J]. J Bacteriol, 2011, 193(1): 177-189.
doi: 10.1128/JB.00895-10 pmid: 20971913 |
| [88] |
Jacob-Dubuisson F, Guérin J, Baelen S, et al. Two-partner secretion: as simple as it sounds?[J]. Res Microbiol, 2013, 164(6): 583-595.
doi: 10.1016/j.resmic.2013.03.009 pmid: 23542425 |
| [89] | Sun YY, Chi H, Sun L. Pseudomonas fluorescens filamentous hemagglutinin, an iron-regulated protein, is an important virulence factor that modulates bacterial pathogenicity[J]. Front Microbiol, 2016, 7: 1320. |
| [90] | Rudzite M, Subramoni S, Endres RG, et al. Effectiveness of Pseu-domonas aeruginosa type VI secretion system relies on toxin potency and type IV pili-dependent interaction[J]. PLoS Pathog, 2023, 19(5): e1011428. |
| [91] |
Lin JS, Xu L, Yang JS, et al. Beyond dueling: roles of the type VI secretion system in microbiome modulation, pathogenesis and stress resistance[J]. Stress Biol, 2021, 1(1): 11.
doi: 10.1007/s44154-021-00008-z pmid: 37676535 |
| [92] | Blanco-Romero E, Garrido-Sanz D, Durán D, et al. Role of extracellular matrix components in biofilm formation and adaptation of Pseudomonas ogarae F113 to the rhizosphere environment[J]. Front Microbiol, 2024, 15: 1341728. |
| [93] | Singh RP, Kumari K. Bacterial type VI secretion system(T6SS): an evolved molecular weapon with diverse functionality[J]. Biotechnol Lett, 2023, 45(3): 309-331. |
| [94] | Bernal P, Civantos C, Pacheco-Sánchez D, et al. Transcriptional organization and regulation of the Pseudomonas putida K1 type VI secretion system gene cluster[J]. Microbiology(Reading), 2023, 169(1): 001295. |
| [95] | Chen WZ, Zhang Y, Zhang YF, et al. CRISPR/Cas9-based genome dditing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species[J]. iScience, 2018, 6: 222-231. |
| [1] | WANG Cai-hong, JIANG Meng-yuan, SHAO Yu-han. Identification and Functional Study of T6SS Effector Protein PA0423 of Pseudomonas aeruginosa [J]. Biotechnology Bulletin, 2024, 40(4): 297-305. |
| [2] | ZHU Ping, DU Li-jie, MENG Kun, XUE Juan, YANG Jin, LI Shan. Research Progress on the Effects of T3SS Effectors on Apoptosis and Pyroptosis of Host Cells [J]. Biotechnology Bulletin, 2019, 35(4): 178-187. |
| [3] | FAN Su-su, TIAN Fang, HE Chen-yang. Regulation and Expression of Genes Encoding the Type III Secretion System in Xanthomonas oryzae pv. oryzae [J]. Biotechnology Bulletin, 2018, 34(2): 38-44. |
| [4] | GAO Ran-ran, LIU Qian, SUN Wen-liang, SUN Zhi-yong, LIU Hao, TIAN Chao-guang. Construction of a Recombinant Protein Expression System in Neurospora crassa [J]. Biotechnology Bulletin, 2016, 32(7): 160-169. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||