








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (5): 52-61.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1033
Previous Articles Next Articles
REN Zhu-ping( ), YANG Tai-ran, LEI Yuan-san, JIN Liu-fei, CUI Gu-zhen, TIAN Yi-ming(
), YANG Tai-ran, LEI Yuan-san, JIN Liu-fei, CUI Gu-zhen, TIAN Yi-ming( ), CHEN Zheng-hong(
), CHEN Zheng-hong( )
)
Received:2024-10-22
Online:2025-05-26
Published:2025-06-05
Contact:
TIAN Yi-ming, CHEN Zheng-hong
E-mail:rzp15685902672@163.com;tianym@gmc.edu.cn;chenzhenghong@gmc.edu.cn
REN Zhu-ping, YANG Tai-ran, LEI Yuan-san, JIN Liu-fei, CUI Gu-zhen, TIAN Yi-ming, CHEN Zheng-hong. Construction of HIEC6-dCas9-SAM Transgenic Cell Line with Highly-efficient CRISPR Synergistic Activation Properties[J]. Biotechnology Bulletin, 2025, 41(5): 52-61.
| 引物 Primer | 序列 Sequence (5′-3′) | 注释 Note |
|---|---|---|
| sgAPN-F | sgAPN慢病毒载体引物 | |
| sgAPN-R | ||
| sgSLC35A1-F | sgSLC35A1慢病毒载体引物 | |
| sgSLC35A1-R | ||
| APN-F | TTCAACATCACGCTTATCCACC | qPCR引物 |
| APN-R | AGTCGAACTCACTGACAATGAAG | |
| SLC35A1-F | CAACCACAGCCGTGTGTATCA | qPCR引物 |
| SLC35A1-R | TGCTAAGAGCTAGGAAAGCCAT | |
| Actin-F | CATGTACGTTGCTATCCAGGC | qPCR引物 |
| Actin-R | CTCCTTAATGTCACGCACGAT |
Table 1 Sequences information
| 引物 Primer | 序列 Sequence (5′-3′) | 注释 Note |
|---|---|---|
| sgAPN-F | sgAPN慢病毒载体引物 | |
| sgAPN-R | ||
| sgSLC35A1-F | sgSLC35A1慢病毒载体引物 | |
| sgSLC35A1-R | ||
| APN-F | TTCAACATCACGCTTATCCACC | qPCR引物 |
| APN-R | AGTCGAACTCACTGACAATGAAG | |
| SLC35A1-F | CAACCACAGCCGTGTGTATCA | qPCR引物 |
| SLC35A1-R | TGCTAAGAGCTAGGAAAGCCAT | |
| Actin-F | CATGTACGTTGCTATCCAGGC | qPCR引物 |
| Actin-R | CTCCTTAATGTCACGCACGAT |
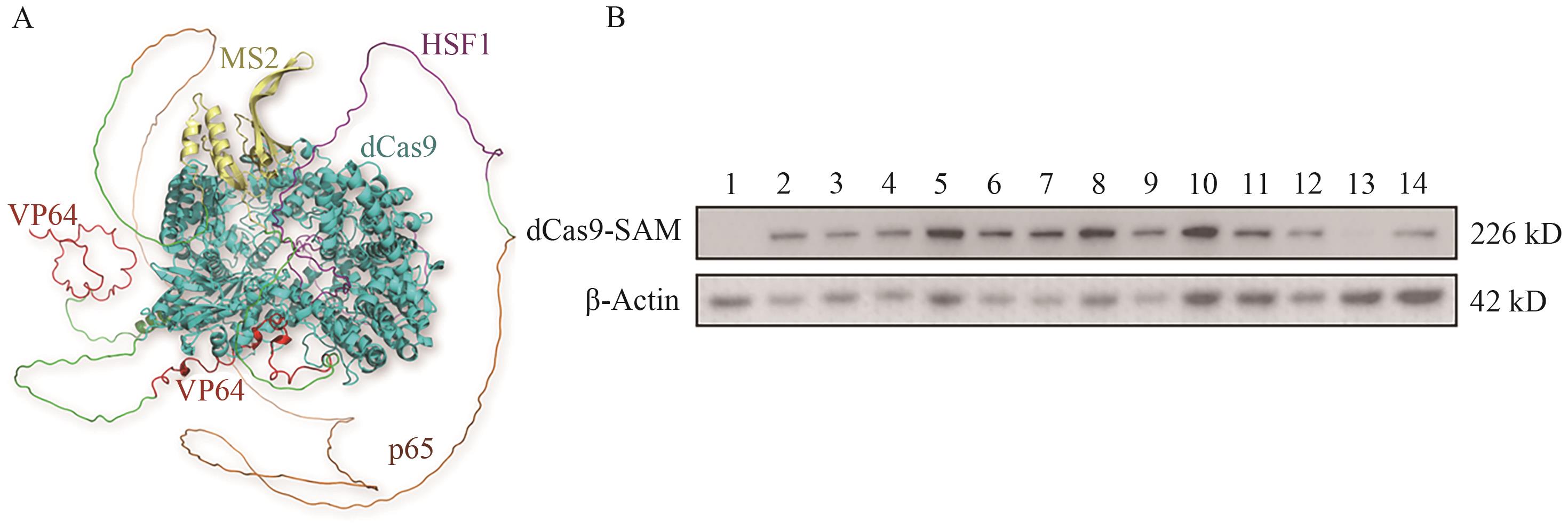
Fig. 1 Identification of dCas9-SAM protein expression in HIEC6 positive polyclonal cells via Western blotA: Diagram of dCas9-SAM protein structure simulation (Cyans: dCas9. Red: VP64. Yellow: MS2. Brown: p65. Purple: HSF1). B: Western blot identification of dCas9-SAM protein expression in positive cells (1: Control of HIEC6 cells; 2-14: HIEC6-dCas9-SAM positive cells)

Fig. 4 CRISPR activation efficiency of HIEC6-dCas9-SAM cells detected by fluorescent reporter systemA: Schematic diagram of CRISPRa fluorescent reporter plasmid and control plasmid (TRE: TetO response element.TetO gRNA: Targeting TRE to recruit dCas9-SAM and drive GFP expression. Empty gRNA: Blank control gRNA). B: Observation of GFP protein expression by fluorescence microscope (Empty: control plasmid. CRISPRa Reporter: CRISPR activation reporter plasmid). C: Detection of activation efficiency by flow cytometry (sgRNA expression/BFP: report plasmid and control plasmid transduced into cells, expressing blue fluorescent protein. GFP: TetO gRNA targeted TRE recruits dCas9-SAM for transcriptional activation and expression of green fluorescent protein)
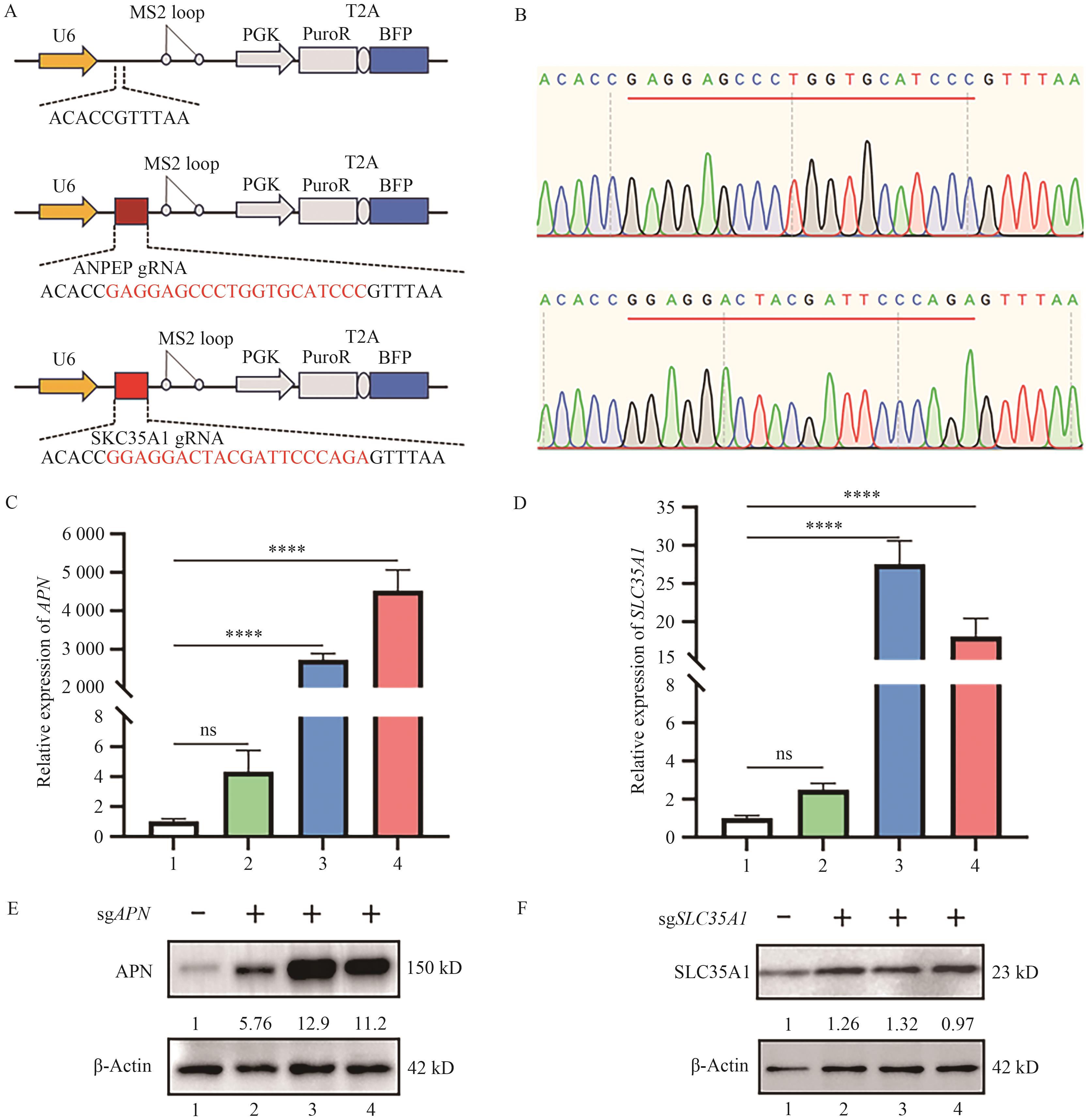
Fig. 5 qRCR and Western blot detection of transcriptional activation of target genes in HIEC6-dCas9-SAM cellsA: Schematics of specific sgRNA lentiviral vector. B: Sequencing result of sgAPN and sgSLC35A1 lentiviral vector. C: Relative expression of APN gene detected by qPCR. D: Relative expression of SLC35A1 gene detected by qPCR (****P<0.000 1 indicates that the relative expression of gene was significantly different between the two groups in one-way ANOVA). E: APN protein expression after CRISPR activation by Western blot. F: SLC35A1 protein expression after CRISPR activation by Western blot (1: Blank;2: HIEC6 cell; 3: 1# HIEC6-dCas9-SAM cell; 4: 4# HIEC6-dCas9-SAM cell)
| 1 | Cong L, Ann Ran F, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems [J]. Science, 2013, 339(6121): 819-823. |
| 2 | Jiang FG, Doudna JA. CRISPR-Cas9 structures and mechanisms [J]. Annu Rev Biophys, 2017, 46: 505-529. |
| 3 | Shields RC, Walker AR, Maricic N, et al. Repurposing the Streptococcus mutans CRISPR-Cas9 system to understand essential gene function [J]. PLoS Pathog, 2020, 16(3): e1008344. |
| 4 | Zhu YM. Advances in CRISPR/Cas9 [J]. BioMed Res Int, 2022, 2022(1): 9978571. |
| 5 | Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation [J]. Cell, 2014, 159(3): 647-661. |
| 6 | Chavez A, Tuttle M, Pruitt BW, et al. Comparison of Cas9 activators in multiple species [J]. Nat Methods, 2016, 13(7): 563-567. |
| 7 | Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex [J]. Nature, 2015, 517(7536): 583-588. |
| 8 | Heidersbach AJ, Dorighi KM, Gomez JA, et al. A versatile, high-efficiency platform for CRISPR-based gene activation [J]. Nat Commun, 2023, 14(1): 902. |
| 9 | Han K, Jeng EE, Hess GT, et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions [J]. Nat Biotechnol, 2017, 35(5): 463-474. |
| 10 | Pan JM, Zhang M, Dong LQ, et al. Genome-Scale CRISPR screen identifies LAPTM5 driving lenvatinib resistance in hepatocellular carcinoma [J]. Autophagy, 2023, 19(4): 1184-1198. |
| 11 | Feng F, Zhu YK, Ma YL, et al. A CRISPR activation screen identifies genes that enhance SARS-CoV-2 infection [J]. Protein Cell, 2022, 14(1): 64-68. |
| 12 | Awwad SW, Serrano-Benitez A, Thomas JC, et al. Revolutionizing DNA repair research and cancer therapy with CRISPR-Cas screens [J]. Nat Rev Mol Cell Biol, 2023, 24(7): 477-494. |
| 13 | Yang LP, Sheets TP, Feng Y, et al. Uncovering receptor-ligand interactions using a high-avidity CRISPR activation screening platform [J]. Sci Adv, 2024, 10(7): eadj2445. |
| 14 | Danziger O, Patel RS, DeGrace EJ, et al. Inducible CRISPR activation screen for interferon-stimulated genes identifies OAS1 as a SARS-CoV-2 restriction factor [J]. PLoS Pathog, 2022, 18(4): e1010464. |
| 15 | Rahman MM, Tollefsbol TO. Targeting cancer epigenetics with CRISPR-dCAS9: principles and prospects [J]. Methods, 2021, 187: 77-91. |
| 16 | Beaulieu JF, Ménard D. Isolation, characterization, and culture of normal human intestinal crypt and villus cells [J]. Methods Mol Biol, 2012, 806: 157-173. |
| 17 | Pászti-Gere E, Pomothy J, Jerzsele Á, et al. Exposure of human intestinal epithelial cells and primary human hepatocytes to trypsin-like serine protease inhibitors with potential antiviral effect [J]. J Enzyme Inhib Med Chem, 2021, 36(1): 659-668. |
| 18 | Perreault N, Beaulieu JF. Use of the dissociating enzyme thermolysin to generate viable human normal intestinal epithelial cell cultures [J]. Exp Cell Res, 1996, 224(2): 354-364. |
| 19 | Pageot LP, Perreault N, Basora N, et al. Human cell models to study small intestinal functions: recapitulation of the crypt-villus axis [J]. Microsc Res Tech, 2000, 49(4): 394-406. |
| 20 | Tremblay E, Auclair J, Delvin E, et al. Gene expression profiles of normal proliferating and differentiating human intestinal epithelial cells: a comparison with the Caco-2 cell model [J]. J Cell Biochem, 2006, 99(4): 1175-1186. |
| 21 | Guezguez A, Paré F, Benoit YD, et al. Modulation of stemness in a human normal intestinal epithelial crypt cell line by activation of the WNT signaling pathway [J]. Exp Cell Res, 2014, 322(2): 355-364. |
| 22 | Juntarachot N, Sunpaweravong S, Kaewdech A, et al. Characterization of adhesion, anti-adhesion, co-aggregation, and hydrophobicity of Helicobacter pylori and probiotic strains [J]. J Taibah Univ Med Sci, 2023, 18(5): 1048-1054. |
| 23 | Jiang YH, Zhang GQ, Li LT, et al. Transcriptomic analysis of PDCoV-infected HIEC-6 cells and enrichment pathways PI3K-Akt and P38 MAPK [J]. Viruses, 2024, 16(4): 579. |
| 24 | Jiang YH, Zhang GQ, Li LT, et al. A novel host restriction factor MRPS6 mediates the inhibition of PDCoV infection in HIEC-6 cells [J]. Front Immunol, 2024, 15: 1381026. |
| 25 | Joung J, Kirchgatterer PC, Singh A, et al. CRISPR activation screen identifies BCL-2 proteins and B3GNT2 as drivers of cancer resistance to T cell-mediated cytotoxicity [J]. Nat Commun, 2022, 13(1): 1606. |
| 26 | 巩琦凡. 基于CRISPRa胃癌耐药相关基因筛选及功能研究 [D]. 保定: 河北大学, 2023. |
| Gong QF. Screening and functional study of drug resistance-related genes in gastric cancer based on CRISPRa [D]. Baoding: Hebei University, 2023. | |
| 27 | Zhu SY, Liu Y, Zhou Z, et al. Genome-wide CRISPR activation screen identifies candidate receptors for SARS-CoV-2 entry [J]. Sci China Life Sci, 2022, 65(4): 701-717. |
| 28 | Clark T, Waller MA, Loo L, et al. CRISPR activation screens: navigating technologies and applications [J]. Trends Biotechnol, 2024, 42(8): 1017-1034. |
| 29 | Sastry L, Xu Y, Duffy L, et al. Product-enhanced reverse transcriptase assay for replication-competent retrovirus and lentivirus detection [J]. Hum Gene Ther, 2005, 16(10): 1227-1236. |
| 30 | Sandoval-Villegas N, Nurieva W, Amberger M, et al. Contemporary transposon tools: a review and guide through mechanisms and applications of Sleeping beauty, piggyBac and Tol2 for genome engineering [J]. Int J Mol Sci, 2021, 22(10): 5084. |
| 31 | Rostovskaya M, Fu J, Obst M, et al. Transposon-mediated BAC transgenesis in human ES cells [J]. Nucleic Acids Res, 2012, 40(19): e150. |
| 32 | Eckermann KN, Ahmed HMM, KaramiNejadRanjbar M, et al. Hyperactive piggyBac transposase improves transformation efficiency in diverse insect species [J]. Insect Biochem Mol Biol, 2018, 98: 16-24. |
| 33 | Vargas JE, Chicaybam L, Stein RT, et al. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives [J]. J Transl Med, 2016, 14(1): 288. |
| 34 | Chong ZS, Ohnishi S, Yusa K, et al. Pooled extracellular receptor-ligand interaction screening using CRISPR activation [J]. Genome Biol, 2018, 19(1): 205. |
| 35 | Zhou J, Wan J, Gao XW, et al. Dynamic m(6)a mRNA methylation directs translational control of heat shock response [J]. Nature, 2015, 526(7574): 591-594. |
| 36 | Moreno-Mateos MA, Vejnar CE, Beaudoin JD, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo [J]. Nat Methods, 2015, 12(10): 982-988. |
| [1] | JIN Zhe-dong, LI Hui-yi, BAO Wen-xin, CUI Cai-xia, YUAN Yun-sheng. Establishment and Application of Stable Cell Lines Expressing Mpox Virus Surface Protein A35R and E8L [J]. Biotechnology Bulletin, 2024, 40(10): 315-322. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | HE Shi-jun, WAN Yi-hong, ZHANG Jia-wen, CAI Xiu-chao, LIU Jing-wen, LIU Shu-wen, YAO Xin-gang. Lentiviral CRISPR/Cas9-Mediated PKA C-α Knockout in Pancreatic-β Cell [J]. Biotechnology Bulletin, 2020, 36(3): 102-109. |
| [4] | YE Zhou-jie, WANG Xin-rui. Research Progress of CRISPR System in Translational Medicine [J]. Biotechnology Bulletin, 2020, 36(11): 188-197. |
| [5] | Su Jialin, Que Biao, Zhang Jiqin, Li Jinhui, Wang Min, Ji Weidong. Establishment of a CRISPR/Cas9 Lentiviral System for Knockout Gene AIP1 of Human [J]. Biotechnology Bulletin, 2015, 31(8): 219-224. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||