








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 124-136.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1113
ZHU Li-juan( ), ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li(
), ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li( )
)
Received:2024-11-14
Online:2025-08-26
Published:2025-07-17
Contact:
WANG Yan-li
E-mail:lijuanzhu91@163.com;yanliwang0720@163.com
ZHU Li-juan, ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li. Mining the Core Genes Being Tolerant to Cadmium in Wild Soybean by WGCNA[J]. Biotechnology Bulletin, 2025, 41(8): 124-136.
性状 Trait | 全距 Range | 最小值 Min | 最大值 Max | 平均值 Average | 标准差 Standard deviation |
|---|---|---|---|---|---|
| SDW-CK1 (g) | 0.090 | 0.006 | 0.096 | 0.017 | 0.008 |
| SDW-Cd1 (g) | 0.049 | 0.001 | 0.050 | 0.013 | 0.005 |
| RDW-CK1 (g) | 0.043 | 0.002 | 0.045 | 0.007 | 0.005 |
| RDW-Cd1 (g) | 0.029 | 0.001 | 0.030 | 0.005 | 0.003 |
| RSR-CK1 | 1.265 | 0.149 | 1.414 | 0.373 | 0.151 |
| RSR-Cd1 | 2.037 | 0.077 | 2.114 | 0.404 | 0.225 |
| CTC-SDW1 | 1.643 | 0.199 | 1.842 | 0.802 | 0.212 |
| CTC-RDW1 | 2.363 | 0.146 | 2.509 | 0.848 | 0.315 |
| CTC-RSR1 | 3.332 | 0.165 | 3.497 | 1.122 | 0.467 |
| SDW-CK2 (g) | 0.082 | 0.007 | 0.089 | 0.016 | 0.008 |
| SDW-Cd2 (g) | 0.059 | 0.002 | 0.061 | 0.014 | 0.007 |
| RDW-CK2 (g) | 0.056 | 0.002 | 0.058 | 0.006 | 0.005 |
| RDW-Cd2 (g) | 0.031 | 0.001 | 0.032 | 0.005 | 0.003 |
| RSR-CK2 | 1.411 | 0.158 | 1.569 | 0.364 | 0.167 |
| RSR-Cd2 | 1.180 | 0.062 | 1.242 | 0.365 | 0.165 |
| CTC-SDW2 | 3.570 | 0.166 | 3.736 | 0.877 | 0.438 |
| CTC-RDW2 | 2.500 | 0.014 | 2.514 | 0.856 | 0.342 |
| CTC-RSR2 | 2.824 | 0.039 | 2.863 | 1.068 | 0.428 |
| D1 | 1.05 | 0.14 | 1.19 | 0.459 | 0.147 |
| D2 | 1.14 | 0.02 | 1.16 | 0.512 | 0.181 |
Table 1 Description analysis of wild soybean seedlings under Cd treatment
性状 Trait | 全距 Range | 最小值 Min | 最大值 Max | 平均值 Average | 标准差 Standard deviation |
|---|---|---|---|---|---|
| SDW-CK1 (g) | 0.090 | 0.006 | 0.096 | 0.017 | 0.008 |
| SDW-Cd1 (g) | 0.049 | 0.001 | 0.050 | 0.013 | 0.005 |
| RDW-CK1 (g) | 0.043 | 0.002 | 0.045 | 0.007 | 0.005 |
| RDW-Cd1 (g) | 0.029 | 0.001 | 0.030 | 0.005 | 0.003 |
| RSR-CK1 | 1.265 | 0.149 | 1.414 | 0.373 | 0.151 |
| RSR-Cd1 | 2.037 | 0.077 | 2.114 | 0.404 | 0.225 |
| CTC-SDW1 | 1.643 | 0.199 | 1.842 | 0.802 | 0.212 |
| CTC-RDW1 | 2.363 | 0.146 | 2.509 | 0.848 | 0.315 |
| CTC-RSR1 | 3.332 | 0.165 | 3.497 | 1.122 | 0.467 |
| SDW-CK2 (g) | 0.082 | 0.007 | 0.089 | 0.016 | 0.008 |
| SDW-Cd2 (g) | 0.059 | 0.002 | 0.061 | 0.014 | 0.007 |
| RDW-CK2 (g) | 0.056 | 0.002 | 0.058 | 0.006 | 0.005 |
| RDW-Cd2 (g) | 0.031 | 0.001 | 0.032 | 0.005 | 0.003 |
| RSR-CK2 | 1.411 | 0.158 | 1.569 | 0.364 | 0.167 |
| RSR-Cd2 | 1.180 | 0.062 | 1.242 | 0.365 | 0.165 |
| CTC-SDW2 | 3.570 | 0.166 | 3.736 | 0.877 | 0.438 |
| CTC-RDW2 | 2.500 | 0.014 | 2.514 | 0.856 | 0.342 |
| CTC-RSR2 | 2.824 | 0.039 | 2.863 | 1.068 | 0.428 |
| D1 | 1.05 | 0.14 | 1.19 | 0.459 | 0.147 |
| D2 | 1.14 | 0.02 | 1.16 | 0.512 | 0.181 |
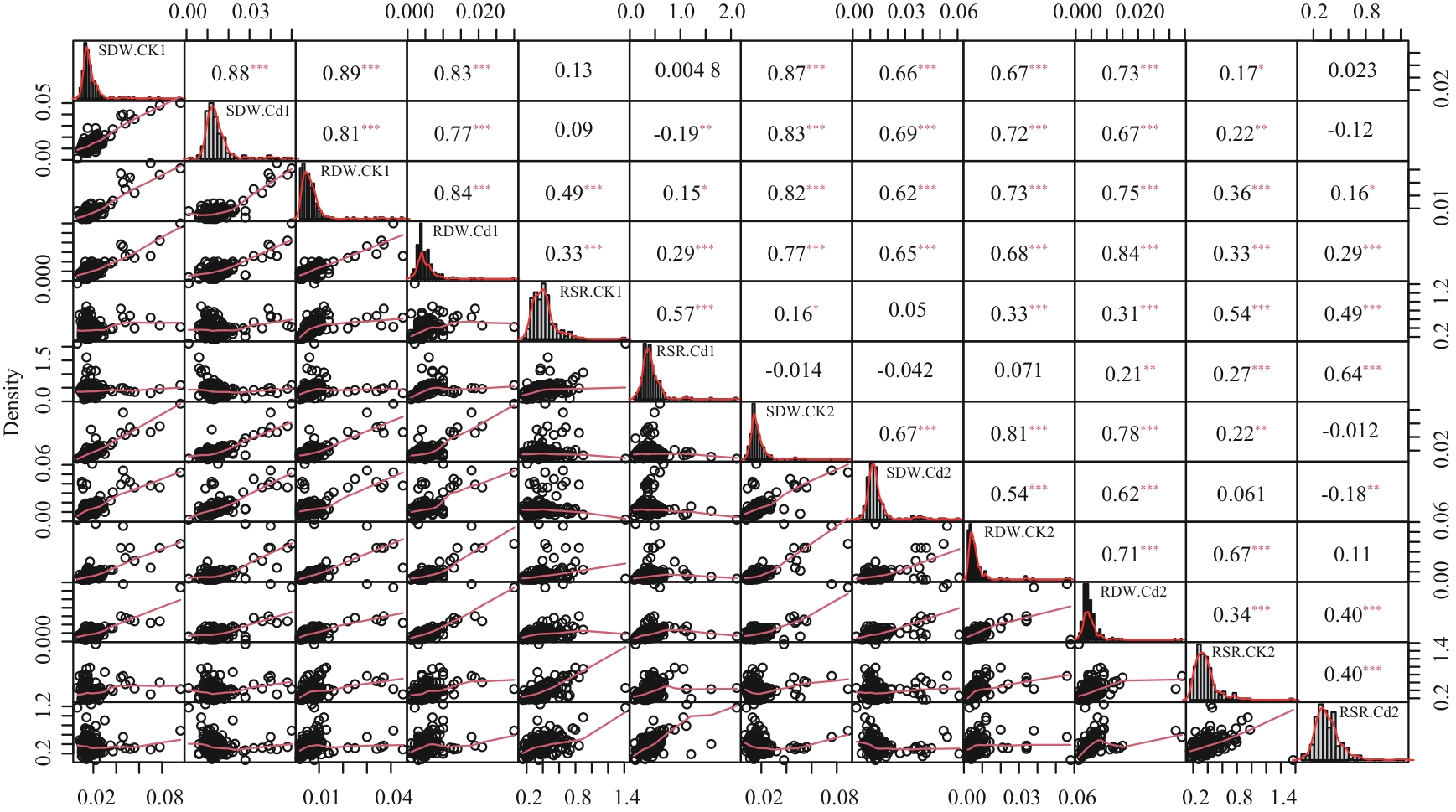
Fig. 1 Correlation analysis of dry weight of wild soybean seedlings under two repetitions*, ** and *** indicate extremely significant correlaton at 0.05, 0.01 and 0.001 levels, respectively

Fig. 3 Analysis of transcriptome sequencing dataA: PCA analysis. B: Correlation test among samples. C: Heat map of DEGs. R0, R24, R48, S0, S24, and S48 indicate 0, 24, and 48 h under Cd stress in R and S. The same below

Fig. 4 Analysis of DEGs of R and S in wild soybean under cadmium stressA: Number of DEGs. B: Venn diagram of DEGs in R at different time. C: Venn diagram of DEGs in S at different time. D: Venn diagram of DEGs in R and S at 24 h under treatment. E: Venn diagram of DEGs in R and S at 48 h under treatment

Fig. 5 GO enrichment analysisA, B: GO enrichment analysis of DEGs in R at 24 h (A) and 48 h (B). C, D: GO enrichment analysis of DEGs in S at 24 h (C) and 48 h (D)
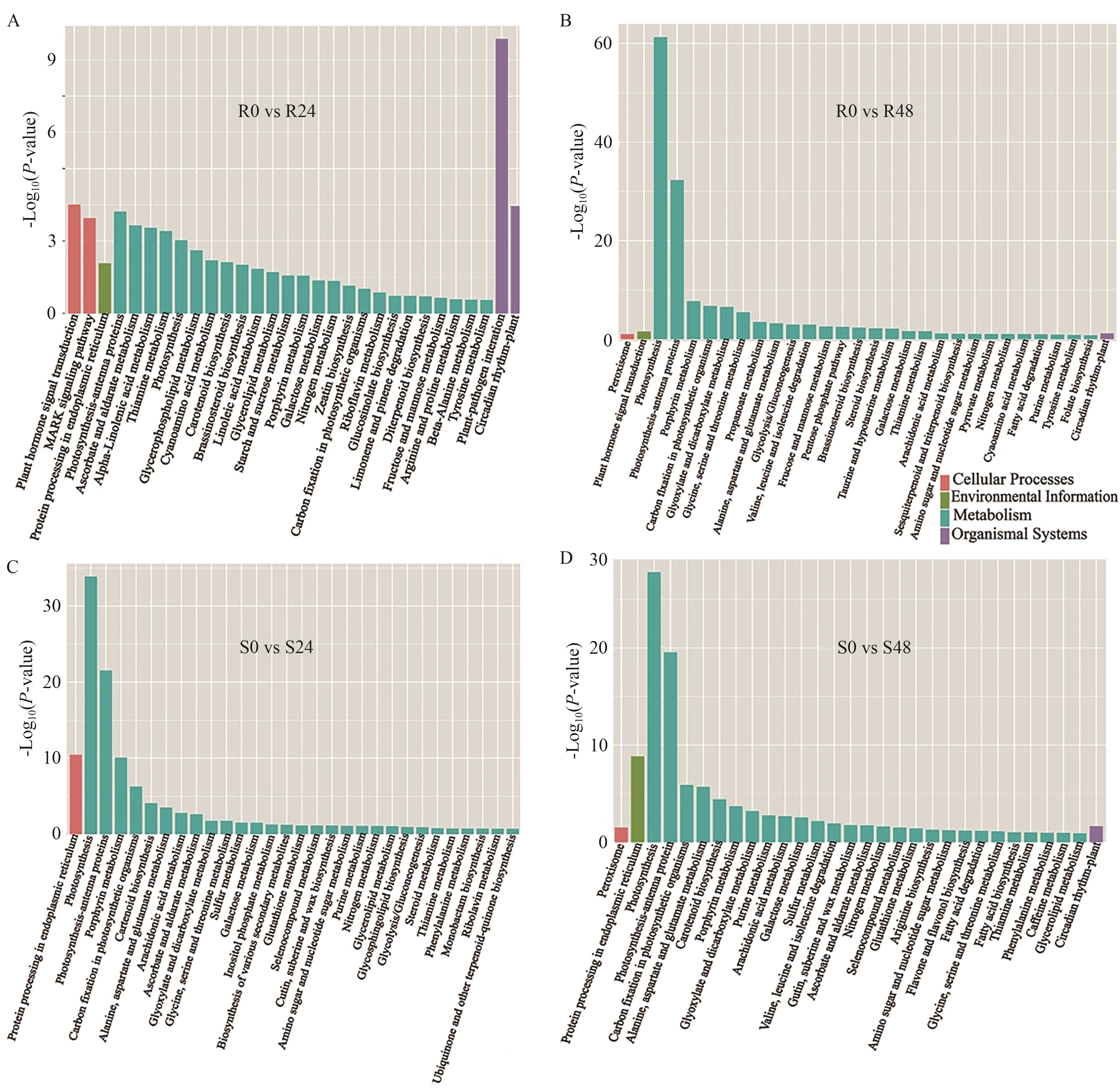
Fig. 6 KEGG enrichment analysiA, B: KEGG enrichment analysis of DEGs in R at 24 h (A) and 48 h (B) under stress. C, D: KEGG enrichment analysis of DEGs in S at 24 h (C) and 48 h (D) under stress

Fig. 7 WGCNA enrichment analysis of DEGs in wild soybean under Cd stressA: Co-expression module division. B: Gene module eigenvalue connectivity heat map. C: Blue module co-expresses the regulatory network. D: Turquoise module co-expresses the regulatory network
关键基因 Gene ID | 基因功能注释 Gene function | 同源基因 Orthologs |
|---|---|---|
| LOC114375809 | Glutamine synthetase leaf isozyme, chloroplastic-like | Glyma 13G210800v4 |
| LOC114376496 | Protein DETOXIFICATION 42-like | Glyma 13G339800v4 |
| LOC114377985 | Stromal 70 kD heat shock-related protein, chloroplastic-like | Glyma 12G166200v4 |
| LOC114387053 | Phosphoglycerate kinase, cytosolic-like | Glyma 15G261900v4 |
| LOC114388638 | Heat shock 70 kD protein 14-like | Glyma 15G014400v4 |
| LOC114399512 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 19G139300v4 |
| LOC114406715 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 03G137000v4 |
| LOC114412091 | FAM10 family protein At4g22670-like | Glyma 05G051600v4 |
Table 2 Functional annotation of 8 WGCNA hub genes
关键基因 Gene ID | 基因功能注释 Gene function | 同源基因 Orthologs |
|---|---|---|
| LOC114375809 | Glutamine synthetase leaf isozyme, chloroplastic-like | Glyma 13G210800v4 |
| LOC114376496 | Protein DETOXIFICATION 42-like | Glyma 13G339800v4 |
| LOC114377985 | Stromal 70 kD heat shock-related protein, chloroplastic-like | Glyma 12G166200v4 |
| LOC114387053 | Phosphoglycerate kinase, cytosolic-like | Glyma 15G261900v4 |
| LOC114388638 | Heat shock 70 kD protein 14-like | Glyma 15G014400v4 |
| LOC114399512 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 19G139300v4 |
| LOC114406715 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 03G137000v4 |
| LOC114412091 | FAM10 family protein At4g22670-like | Glyma 05G051600v4 |
| [1] | Mubeen S, Ni WJ, He CT, et al. Agricultural strategies to reduce cadmium accumulation in crops for food safety [J]. Agriculture, 2023, 13(2): 471. |
| [2] | Yan HL, Hezifan Z, Hao SN, et al. Cadmium contamination in food crops: risk assessment and control in smart age [J]. Crit Rev Environ Sci Technol, 2023, 53(18): 1643-1661. |
| [3] | Genchi G, Sinicropi MS, Lauria G, et al. The effects of cadmium toxicity [J]. Int J Environ Res Public Health, 2020, 17(11): 3782. |
| [4] | Finger-Teixeira A, de Lourdes Lucio Ferrarese M, Ricardo Soares A, et al. Cadmium-induced lignification restricts soybean root growth [J]. Ecotoxicol Environ Saf, 2010, 73(8): 1959-1964. |
| [5] | He SL, Wang YS, Li DZ, et al. Environmental and historical determinants of patterns of genetic differentiation in wild soybean (Glycine soja sieb. et zucc) [J]. Sci Rep, 2016, 6: 22795. |
| [6] | Wang KJ, Li F, Ali Cheema A. Studies on the distribution of wild soybean (Glycine soja) in China [J]. Pak J Biol Sci, 2001, 4(2): 149-155. |
| [7] | 李明霞. 盐和低氮胁迫下栽培大豆和野大豆适应性比较研究 [D]. 长春: 东北师范大学, 2020. |
| Li MX. Comparative study on adaptability of cultivated soybean and wild soybean under salt and low nitrogen stress [D]. Changchun: Northeast Normal University, 2020. | |
| [8] | Liu DP, Li MX, Liu Y, et al. Integration of the metabolome and transcriptome reveals the resistance mechanism to low nitrogen in wild soybean seedling roots [J]. Environ Exp Bot, 2020, 175: 104043. |
| [9] | 赵忠娟, 魏艳丽, 李哲, 等. 野大豆与栽培大豆愈伤组织耐盐性比较 [J]. 山东科学, 2015, 28(1): 102-108. |
| Zhao ZJ, Wei YL, Li Z, et al. Salinity tolerance comparison for the calli of wild and cultivated soybeans [J]. Shandong Sci, 2015, 28(1): 102-108. | |
| [10] | 符辉. 干旱胁迫下野大豆(Glycine soja sieb.et zucc.)和大豆(Glycine max L.)幼苗叶片代谢组学比较研究 [D]. 长春: 东北师范大学, 2020. |
| Fu H. Comparative study on leaf metabonomics of Glycine soja sieb.et zucc. and Glycine max L. seedlings under drought stress [D]. Changchun: Northeast Normal University, 2020. | |
| [11] | Gao Y, Tao B, Qiu LJ, et al. Role of physiological mechanisms and EPSPS gene expression in glyphosate resistance in wild soybeans (Glycine soja) [J]. Pestic Biochem Physiol, 2014, 109: 6-11. |
| [12] | 袁翠平, 齐广勋, 李玉秋, 等. 野生大豆抗胞囊线虫QTL定位 [J]. 中国油料作物学报, 2019, 41(6): 887-893. |
| Yuan CP, Qi GX, Li YQ, et al. QTL mapping for resistance to soybean cyst nematode in wild soybean [J]. Chin J Oil Crop Sci, 2019, 41(6): 887-893. | |
| [13] | 陈爱国, 王岩, 孟未来, 等. 不同原生境来源野生大豆抗花叶病毒(SMV)综合评价及聚类分析 [J]. 辽宁农业科学, 2020(1): 7-13. |
| Chen AG, Wang Y, Meng WL, et al. Evaluation, cluster analysis for Glycine soja of different habitats resistant to soybean mosaic virus (SMV) [J]. Liaoning Agric Sci, 2020(1): 7-13. | |
| [14] | 陈珊宇, 王大刚, 郑桂杰, 等. 野生大豆对大豆花叶病毒株系SC13的抗性遗传和基因定位 [J]. 植物遗传资源学报, 2020, 21(1): 139-145. |
| Chen SY, Wang DG, Zheng GJ, et al. Inheritance and gene mapping of resistance to soybean mosaic virus strain SC13 in soybean [Glycine soja sieb. & zucc.] [J]. J Plant Genet Resour, 2020, 21(1): 139-145. | |
| [15] | Arao T, Ae N, Sugiyama MT, et al. Genotypic differences in cadmium uptake and distribution in soybeans [J]. Plant Soil, 2003, 251(2): 247-253. |
| [16] | 赵毅, 于翠梅, 杨柳, 等. 野生大豆和不同栽培大豆品种在镉胁迫下种子萌发及幼苗生长的差异 [J]. 大豆科学, 2019, 38(2): 267-273. |
| Zhao Y, Yu CM, Yang L, et al. Differences of cadmium stress on seed germination and seedling growth in the wild soybean and cultivated soybeans [J]. Soybean Sci, 2019, 38(2): 267-273. | |
| [17] | 冯君, 赵毅, 高婷, 等. 野生和栽培大豆对镉胁迫的响应差异分析 [J]. 大豆科学, 2018, 37(5): 756-761. |
| Feng J, Zhao Y, Gao T, et al. The difference of responses to the cadmium stress between a wild soybean and a cultivated soybean [J]. Soybean Sci, 2018, 37(5): 756-761. | |
| [18] | Yu HW, Yang ZM, Wang JF, et al. Identification of key genes and metabolites involved in meat quality performance in Qinchuan cattle by WGCNA [J]. J Integr Agric, 2024, 23(11): 3923-3937. |
| [19] | 夏雪岩, 崔纪菡, 黄玫红, 等. 谷子苗期氮高效转录组分析与基因挖掘 [J]. 中国农业科技导报, 2024, 26(10): 41-57. |
| Xia XY, Cui JH, Huang MH, et al. Analysis of high-efficiency transcriptome of nitrogen in millet seedlings and gene mining [J]. J Agric Sci Technol, 2024, 26(10): 41-57. | |
| [20] | 陈晓涓, 王海菊, 王富敏, 等. 基于WGCNA鉴定全缘叶绿绒蒿类黄酮合成途径关键基因 [J]. 中国农业科学, 2024, 57(15): 3053-3070. |
| Chen XJ, Wang HJ, Wang FM, et al. Identification of key genes in the flavonoid synthesis pathway of Meconopsis integrifolia based on WGCNA [J]. Sci Agric Sin, 2024, 57(15): 3053-3070. | |
| [21] | 张会, 王越越, 赵波, 等. 基于WGCNA的谷子苗期冷胁迫应答基因网络构建与核心因子发掘 [J]. 中国农业科技导报, 2023, 25(10): 22-34. |
| Zhang H, Wang YY, Zhao B, et al. Identification of co-expression genes related to cold stress in foxtail millet by WGCNA [J]. J Agric Sci Technol, 2023, 25(10): 22-34. | |
| [22] | 周雨青, 杨永飞, 葛常伟, 等. 基于WGCNA的棉花子叶抗冷相关共表达模块鉴定 [J]. 中国农业科技导报, 2022, 24(4): 52-62. |
| Zhou YQ, Yang YF, Ge CW, et al. Identification of cold-related co-expression modules in cotton Cotyledon by WGCNA [J]. J Agric Sci Technol, 2022, 24(4): 52-62. | |
| [23] | 王世瑶. 低氮胁迫下不同生态型野大豆(Glycine soja)新老叶片光合特性,离子平衡和氮代谢的研究 [D]. 长春: 东北师范大学, 2021. |
| Wang SY. Adaptive variations for nitrogen deficiency in photosynthetic characteristics, ion balance and nitrogen metabolism in young and old leaves of different ecotypes wild soybean[D]. Changchun: Northeast Normal University, 2021. | |
| [24] | Liu XL, Zhang CL, Lamlom SF, et al. Genetic adaptations of soybean to cold stress reveal key insights through transcriptomic analysis [J]. Biology, 2024, 13(11): 856. |
| [25] | Cheng Y, Cheng XQ, Wei K, et al. Comparative transcriptome analysis of salt-tolerant and-sensitive soybean cultivars under salt stress [J]. Int J Mol Sci, 2024, 25(18): 9818. |
| [26] | Li MQ, Li HN, Sun AN, et al. Transcriptome analysis reveals key drought-stress-responsive genes in soybean [J]. Front Genet, 2022, 13: 1060529. |
| [27] | Zheng SL, Qi J, Fu TW, et al. Novel mechanisms of cadmium tolerance and Cd-induced fungal stress in wheat: Transcriptomic and metagenomic insights [J]. Ecotoxicol Environ Saf, 2023, 256: 114842. |
| [28] | 罗玲, 许肖恒, 杨康, 等. 非生物胁迫下植物衰老和热激蛋白响应 [J]. 草业科学, 2020, 37(11): 2320-2333. |
| Luo L, Xu XH, Yang K, et al. Senescence and heat shock protein in plants in response to abiotic stress [J]. Pratacultural Sci, 2020, 37(11): 2320-2333. | |
| [29] | Luo JS, Zhang ZH. Mechanisms of cadmium phytoremediation and detoxification in plants [J]. Crop J, 2021, 9(3): 521-529. |
| [30] | Feng Z, Ji SY, Ping JF, et al. Recent advances in metabolomics for studying heavy metal stress in plants [J]. Trac Trends Anal Chem, 2021, 143: 116402. |
| [31] | Ma SC, Lapin D, Liu L, et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme [J]. Science, 2020, 370(6521): eabe3069. |
| [1] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [2] | GUO Xiu-juan, FENG Yu, WU Rui-xiang, WANG Li-qin, YANG Jian-chun. Transcriptome Analysis of the Effect of Ca 2+ Treatment on the Seed Germination of Flax [J]. Biotechnology Bulletin, 2025, 41(7): 139-149. |
| [3] | LI Xu-juan, LI Chun-jia, LIU Hong-bo, XU Chao-hua, LIN Xiu-qin, LU Xin, LIU Xin-long. Transcriptome Analysis of Axillary Bud Formation and Development in Sugarcane [J]. Biotechnology Bulletin, 2025, 41(3): 202-218. |
| [4] | ZHAO Hai-ping, LIU Lin, WANG Xin-lu, YUE Peng-fei, KONG Wei-jun, WANG Meng. Exploring the Protective Effect of Puerarin on Deoxynivalenol-induced C6 Cell Injury Based on WGCNA [J]. Biotechnology Bulletin, 2024, 40(9): 301-310. |
| [5] | WEN Jie, DU Yuan-xin, WU An-bo, YANG Guang-rong, LU Min, AN Hua-ming, NAN Hong. Identification and Expression Pattern Analysis of Rosa roxburghii SOD Gene Family [J]. Biotechnology Bulletin, 2024, 40(5): 153-166. |
| [6] | LIU Zi-ran, ZHEN Zhen, CHEN Qiang, LI Yue-ying, WANG Ze, PANG Hong-bo. Research Progress in Plant Response to Cd Stress [J]. Biotechnology Bulletin, 2022, 38(6): 13-26. |
| [7] | GUO Ying, YANG Ping, ZHANG Dan-yu, LIU Ying-ying, MA Lian-ju, BU Ning. Screening,Identification and Growth-promoting Effect of Multi-function Rhizosphere Growth-promoting Strain of Wild Soybean [J]. Biotechnology Bulletin, 2018, 34(10): 108-115. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||