








生物技术通报 ›› 2022, Vol. 38 ›› Issue (6): 81-92.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1102
钟辉1,2( ), 刘亚军1,2, 王滨花1,2, 和梦洁1,2, 吴兰1,2(
), 刘亚军1,2, 王滨花1,2, 和梦洁1,2, 吴兰1,2( )
)
收稿日期:2021-08-27
出版日期:2022-06-26
发布日期:2022-07-11
作者简介:钟辉,男,硕士研究生,研究方向:微生物生态学;E-mail: 基金资助:
ZHONG Hui1,2( ), LIU Ya-jun1,2, WANG Bin-hua1,2, HE Meng-jie1,2, WU Lan1,2(
), LIU Ya-jun1,2, WANG Bin-hua1,2, HE Meng-jie1,2, WU Lan1,2( )
)
Received:2021-08-27
Published:2022-06-26
Online:2022-07-11
摘要:
细菌16S rRNA基因扩增测序是当前环境微生物组学研究中应用最为广泛的方法之一。然而,测序序列最小分类单元的划分有多种方式,其对微生物多样性下游分析结果的影响还有待进一步探究。本研究通过提取5组环境样本(森林、农田、湿地土壤、湖泊沉积物和水体)的DNA进行16S rRNA基因扩增测序,对测序结果同时采用5种最小分类单元的划分方式(基于97%、98%、99%和100%序列相似性聚类的OTU以及基于DADA2算法得到的ASV)进行划分,比较分析最小分类单元划分方法对微生物群落多样性、组成、以及其与环境因子关联性分析造成的影响。结果表明,提高分类分辨率,能够获得更高的群落α多样性(Chao1和Shannon)和β多样性(P < 0.05),而相对于按序列相似性聚类的OTU,ASV方法会在一定程度上降低Chao1和PD指数。对于群落组成,分类单元的划分方式主要影响微生物组一些低丰度属(< 0.2%)的占比,而对较高的分类学水平(门水平)组成的影响较小。此外,冗余分析的结果表明,提高分类分辨率水平,能够使得环境因子对微生物群落能够获得更高的解释度,同时也会影响各环境因子对群落组成的解释度排序。总之,本研究明晰了最小分类单元的不同划分方式会对微生物组多样性、组成以及与环境因子的关联性造成的影响,为后续环境微生物组学研究提供了理论指导。
钟辉, 刘亚军, 王滨花, 和梦洁, 吴兰. 分析方法对细菌群落16S rRNA基因扩增测序分析结果的影响[J]. 生物技术通报, 2022, 38(6): 81-92.
ZHONG Hui, LIU Ya-jun, WANG Bin-hua, HE Meng-jie, WU Lan. Effects of Analysis Methods on the Analyzed Results of 16S rRNA Gene Amplicon Sequencing in Bacterial Communities[J]. Biotechnology Bulletin, 2022, 38(6): 81-92.
| 样本 Sample | 样本类型 Sample type | 采样时间 Sampling time | 酸碱度 pH | 总有机碳 Total organic carbon(TOC)/(g·kg-1) | 总氮 Total nitrogen(TN)/(g·kg-1) | 总磷 Total phosphorus(TP)/(g·kg-1) | 氨态氮 NH4+ N/ (mg·kg-1) | 硝态氮 NO3- N/ (mg·kg-1) |
|---|---|---|---|---|---|---|---|---|
| FS1 | 森林土壤 | 2016年8月 | 4.57±0.24 | 48.07±1.17 | 3.98±0.26 | 0.49±0.09 | 9.01±0.64 | 45.14±11.26 |
| FS2 | 森林土壤 | 2016年8月 | 4.64±0.1 | 58.29±9.12 | 3.86±0.62 | 0.68±0.17 | 46.73±4.21 | 12.19±2.08 |
| FS3 | 森林土壤 | 2016年8月 | 4.70±0.12 | 55.17±4.91 | 3.93±0.83 | 0.56±0.11 | 13.24±1.48 | 27.90±3.41 |
| FS4 | 森林土壤 | 2016年8月 | 5.35±0.67 | 22.96±0.86 | 2.68±0.54 | 0.80±0.14 | 9.06±1.05 | 32.48±7.65 |
| WS1 | 湿地土壤 | 2015年1月 | 5.29±0.14 | 7.31±1.68 | 0.92±0.25 | 0.60±0.18 | 2.30±0.27 | 0.71±0.06 |
| WS2 | 湿地土壤 | 2015年1月 | 4.95±0.58 | 6.89±1.78 | 0.97±0.26 | 0.64±0.19 | 2.85±0.42 | 0.42±0.04 |
| WS3 | 湿地土壤 | 2015年1月 | 5.19±0.25 | 8.82±3.33 | 0.98±0.23 | 0.54±0.06 | 2.79±0.33 | 0.75±0.42 |
| WS4 | 湿地土壤 | 2015年1月 | 5.72±0.39 | 6.64±2.81 | 1.02±0.24 | 0.41±0.06 | 2.65±0.47 | 1.07±0.36 |
| CS1 | 农田土壤 | 2017年5月 | 4.71±0.54 | 36.60±4.66 | 3.95±0.15 | 0.45±0.19 | 0.91±0.11 | 2.54±1.18 |
| CS2 | 农田土壤 | 2017年5月 | 5.22±0.02 | 66.34±3.13 | 7.86±1.15 | 0.55±0.09 | 1.05±0.11 | 5.81±0.25 |
| CS3 | 农田土壤 | 2017年5月 | 5.17±0.17 | 41.11±1.62 | 4.13±0.36 | 0.26±0.01 | 0.87±0.12 | 3.37±0.80 |
| CS4 | 农田土壤 | 2017年5月 | 5.06±0.07 | 44.49±2.11 | 4.97±0.13 | 0.52±0.13 | 0.96±0.08 | 6.52±0.32 |
| LS1 | 湖泊沉积物 | 2018年5月 | 6.60±0.35 | 9.78±1.61 | 1.01±0.14 | 0.96±0.24 | 15.72±9.03 | 1.18±0.68 |
| LS2 | 湖泊沉积物 | 2018年5月 | 7.30±0.22 | 9.97±5.50 | 14.40±2.07 | 1.15±0.16 | 22.10±7.40 | 1.75±0.57 |
| LS3 | 湖泊沉积物 | 2018年5月 | 6.63±0.03 | 7.47±6.21 | 6.96±2.41 | 0.71±0.11 | 5.51±1.08 | 1.68±1.84 |
| LS4 | 湖泊沉积物 | 2018年5月 | 8.08±0.08 | 5.32±3.72 | 14.37±8.57 | 0.90±0.22 | 16.83±2.47 | 1.79±1.04 |
表1 土壤和沉积物样本信息表及环境参数
Table1 Information and environmental parameters of soil and sediment samples
| 样本 Sample | 样本类型 Sample type | 采样时间 Sampling time | 酸碱度 pH | 总有机碳 Total organic carbon(TOC)/(g·kg-1) | 总氮 Total nitrogen(TN)/(g·kg-1) | 总磷 Total phosphorus(TP)/(g·kg-1) | 氨态氮 NH4+ N/ (mg·kg-1) | 硝态氮 NO3- N/ (mg·kg-1) |
|---|---|---|---|---|---|---|---|---|
| FS1 | 森林土壤 | 2016年8月 | 4.57±0.24 | 48.07±1.17 | 3.98±0.26 | 0.49±0.09 | 9.01±0.64 | 45.14±11.26 |
| FS2 | 森林土壤 | 2016年8月 | 4.64±0.1 | 58.29±9.12 | 3.86±0.62 | 0.68±0.17 | 46.73±4.21 | 12.19±2.08 |
| FS3 | 森林土壤 | 2016年8月 | 4.70±0.12 | 55.17±4.91 | 3.93±0.83 | 0.56±0.11 | 13.24±1.48 | 27.90±3.41 |
| FS4 | 森林土壤 | 2016年8月 | 5.35±0.67 | 22.96±0.86 | 2.68±0.54 | 0.80±0.14 | 9.06±1.05 | 32.48±7.65 |
| WS1 | 湿地土壤 | 2015年1月 | 5.29±0.14 | 7.31±1.68 | 0.92±0.25 | 0.60±0.18 | 2.30±0.27 | 0.71±0.06 |
| WS2 | 湿地土壤 | 2015年1月 | 4.95±0.58 | 6.89±1.78 | 0.97±0.26 | 0.64±0.19 | 2.85±0.42 | 0.42±0.04 |
| WS3 | 湿地土壤 | 2015年1月 | 5.19±0.25 | 8.82±3.33 | 0.98±0.23 | 0.54±0.06 | 2.79±0.33 | 0.75±0.42 |
| WS4 | 湿地土壤 | 2015年1月 | 5.72±0.39 | 6.64±2.81 | 1.02±0.24 | 0.41±0.06 | 2.65±0.47 | 1.07±0.36 |
| CS1 | 农田土壤 | 2017年5月 | 4.71±0.54 | 36.60±4.66 | 3.95±0.15 | 0.45±0.19 | 0.91±0.11 | 2.54±1.18 |
| CS2 | 农田土壤 | 2017年5月 | 5.22±0.02 | 66.34±3.13 | 7.86±1.15 | 0.55±0.09 | 1.05±0.11 | 5.81±0.25 |
| CS3 | 农田土壤 | 2017年5月 | 5.17±0.17 | 41.11±1.62 | 4.13±0.36 | 0.26±0.01 | 0.87±0.12 | 3.37±0.80 |
| CS4 | 农田土壤 | 2017年5月 | 5.06±0.07 | 44.49±2.11 | 4.97±0.13 | 0.52±0.13 | 0.96±0.08 | 6.52±0.32 |
| LS1 | 湖泊沉积物 | 2018年5月 | 6.60±0.35 | 9.78±1.61 | 1.01±0.14 | 0.96±0.24 | 15.72±9.03 | 1.18±0.68 |
| LS2 | 湖泊沉积物 | 2018年5月 | 7.30±0.22 | 9.97±5.50 | 14.40±2.07 | 1.15±0.16 | 22.10±7.40 | 1.75±0.57 |
| LS3 | 湖泊沉积物 | 2018年5月 | 6.63±0.03 | 7.47±6.21 | 6.96±2.41 | 0.71±0.11 | 5.51±1.08 | 1.68±1.84 |
| LS4 | 湖泊沉积物 | 2018年5月 | 8.08±0.08 | 5.32±3.72 | 14.37±8.57 | 0.90±0.22 | 16.83±2.47 | 1.79±1.04 |
| 样本 Sample | 样本类型 Sample type | 采样时间 Sampling time | 酸碱度 pH | 总有机碳 TOC/(mg·kg-1) | 总氮 TN/(mg·kg-1) | 总磷 TP/(mg·kg-1) | 氨态氮 NH4+ N/(mg·kg-1) | 硝态氮 NO3--N/(mg·kg-1) |
|---|---|---|---|---|---|---|---|---|
| LW1 | 湖泊水体 | 2017年7月 | 7.87±0.50 | 16.39±6.93 | 2.40±0.66 | 0.13±0.03 | 0.26±0.10 | 0.66±0.12 |
| LW2 | 湖泊水体 | 2017年7月 | 7.3±0.16 | 12.40±5.44 | 3.10±0.63 | 0.13±0.02 | 0.14±0.05 | 0.26±0.04 |
| LW3 | 湖泊水体 | 2017年7月 | 6.79±0.41 | 11.09±5.51 | 1.38±0.36 | 0.11±0.04 | 0.34±0.15 | 0.48±0.04 |
| LW4 | 湖泊水体 | 2017年7月 | 7.12±0.13 | 14.06±6.00 | 1.14±0.67 | 0.11±0.04 | 0.25±0.06 | 0.02±0.03 |
表2 水体样本信息表及环境参数
Table2 Water samples information and environmental parameters
| 样本 Sample | 样本类型 Sample type | 采样时间 Sampling time | 酸碱度 pH | 总有机碳 TOC/(mg·kg-1) | 总氮 TN/(mg·kg-1) | 总磷 TP/(mg·kg-1) | 氨态氮 NH4+ N/(mg·kg-1) | 硝态氮 NO3--N/(mg·kg-1) |
|---|---|---|---|---|---|---|---|---|
| LW1 | 湖泊水体 | 2017年7月 | 7.87±0.50 | 16.39±6.93 | 2.40±0.66 | 0.13±0.03 | 0.26±0.10 | 0.66±0.12 |
| LW2 | 湖泊水体 | 2017年7月 | 7.3±0.16 | 12.40±5.44 | 3.10±0.63 | 0.13±0.02 | 0.14±0.05 | 0.26±0.04 |
| LW3 | 湖泊水体 | 2017年7月 | 6.79±0.41 | 11.09±5.51 | 1.38±0.36 | 0.11±0.04 | 0.34±0.15 | 0.48±0.04 |
| LW4 | 湖泊水体 | 2017年7月 | 7.12±0.13 | 14.06±6.00 | 1.14±0.67 | 0.11±0.04 | 0.25±0.06 | 0.02±0.03 |
| 群落生境 Biotope | 多样性指数 Diversity index | 97 OTU | 98 OTU | 99 OTU | 100 OTU | ASV | F | P |
|---|---|---|---|---|---|---|---|---|
| FS | Shannon | 8.89±0.59c | 9.44±0.57c | 10.06±0.53b | 11.35±0.34a | 9.36±0.36c | 3.76 | 0.01 |
| Faith’s phylog-enetic diversity | 200.65±30.09a | 206.76±30.33a | 205.45±31.21a | 189.98±30.05a | 76.57±17.53b | 3.59 | 0.01 | |
| Chao1 | 6 197.73±681.64d | 7 715.24±864.03c | 9 783.86±1121.87b | 15 783.82±1 896.16a | 1 348.3±250.55e | 64.05 | <0.01 | |
| WS | Shannon | 9.15±0.44cd | 9.51±0.40c | 9.93±0.33b | 10.76±0.22a | 9.10±0.25d | 45.32 | <0.01 |
| Faith’s phylog-enetic diversity | 131.82±20.02ab | 135.38±20.39a | 131.70±21.02ab | 112.17±17.56b | 59.95±11.00c | 46.78 | <0.01 | |
| Chao1 | 3 432.91±489.63c | 4 119.94±523.16bc | 4 834.80±605.21b | 6 259.35±1 102.91a | 909.03±166.82d | 272.21 | <0.01 | |
| CS | Shannon | 8.99±0.88b | 9.27±0.93b | 9.60±1.01ab | 10.40±1.00a | 9.42±0.92ab | 3.51 | 0.01 |
| Faith’s phylog-enetic diversity | 180.30±33.33ab | 169.08±31.10ab | 202.30±38.74a | 199.45±36.96a | 159.09±30.77b | 1.1 | 0.37 | |
| Chao1 | 3 672.12±892.42c | 4 271.85±1 029.13bc | 4 993.04±1 280.13b | 9 069.09±1 554.95a | 2 186.44±570.72d | 51.61 | <0.01 | |
| LS | Shannon | 6.82±1.06b | 6.96±1.15b | 7.21±1.24ab | 8.32±1.09a | 7.20±0.90ab | 49.53 | <0.01 |
| Faith’s phylog-enetic diversity | 97.39±35.21a | 97.87±34.88a | 99.98±35.44a | 110.06±32.85a | 83.22±15.54a | 35.66 | <0.01 | |
| Chao1 | 1 429.92±867.84bc | 1 646.30±1 010.35bc | 1 916.62±1 242.16b | 5 343.72±481.48a | 934.21±263.76c | 110.54 | <0.01 | |
| LW | Shannon | 6.71±1.12b | 6.98±1.14b | 7.36±1.10b | 9.04±0.71a | 7.50±0.80b | 10.01 | <0.01 |
| Faith’s phylog-enetic diversity | 79.77±30.70a | 88.65±32.64a | 91.25±32.18a | 85.25±24.93a | 59.01±18.95a | 2.48 | 0.05 | |
| Chao1 | 1 709.26±868.06bc | 2 043.13±914.34bc | 2 674.50±1 066.06b | 8 847.05±1 696.64a | 837.88±332.8c | 108.4 | <0.01 |
表3 细菌群落的α多样性
Table 3 Alpha diversity of bacterial community
| 群落生境 Biotope | 多样性指数 Diversity index | 97 OTU | 98 OTU | 99 OTU | 100 OTU | ASV | F | P |
|---|---|---|---|---|---|---|---|---|
| FS | Shannon | 8.89±0.59c | 9.44±0.57c | 10.06±0.53b | 11.35±0.34a | 9.36±0.36c | 3.76 | 0.01 |
| Faith’s phylog-enetic diversity | 200.65±30.09a | 206.76±30.33a | 205.45±31.21a | 189.98±30.05a | 76.57±17.53b | 3.59 | 0.01 | |
| Chao1 | 6 197.73±681.64d | 7 715.24±864.03c | 9 783.86±1121.87b | 15 783.82±1 896.16a | 1 348.3±250.55e | 64.05 | <0.01 | |
| WS | Shannon | 9.15±0.44cd | 9.51±0.40c | 9.93±0.33b | 10.76±0.22a | 9.10±0.25d | 45.32 | <0.01 |
| Faith’s phylog-enetic diversity | 131.82±20.02ab | 135.38±20.39a | 131.70±21.02ab | 112.17±17.56b | 59.95±11.00c | 46.78 | <0.01 | |
| Chao1 | 3 432.91±489.63c | 4 119.94±523.16bc | 4 834.80±605.21b | 6 259.35±1 102.91a | 909.03±166.82d | 272.21 | <0.01 | |
| CS | Shannon | 8.99±0.88b | 9.27±0.93b | 9.60±1.01ab | 10.40±1.00a | 9.42±0.92ab | 3.51 | 0.01 |
| Faith’s phylog-enetic diversity | 180.30±33.33ab | 169.08±31.10ab | 202.30±38.74a | 199.45±36.96a | 159.09±30.77b | 1.1 | 0.37 | |
| Chao1 | 3 672.12±892.42c | 4 271.85±1 029.13bc | 4 993.04±1 280.13b | 9 069.09±1 554.95a | 2 186.44±570.72d | 51.61 | <0.01 | |
| LS | Shannon | 6.82±1.06b | 6.96±1.15b | 7.21±1.24ab | 8.32±1.09a | 7.20±0.90ab | 49.53 | <0.01 |
| Faith’s phylog-enetic diversity | 97.39±35.21a | 97.87±34.88a | 99.98±35.44a | 110.06±32.85a | 83.22±15.54a | 35.66 | <0.01 | |
| Chao1 | 1 429.92±867.84bc | 1 646.30±1 010.35bc | 1 916.62±1 242.16b | 5 343.72±481.48a | 934.21±263.76c | 110.54 | <0.01 | |
| LW | Shannon | 6.71±1.12b | 6.98±1.14b | 7.36±1.10b | 9.04±0.71a | 7.50±0.80b | 10.01 | <0.01 |
| Faith’s phylog-enetic diversity | 79.77±30.70a | 88.65±32.64a | 91.25±32.18a | 85.25±24.93a | 59.01±18.95a | 2.48 | 0.05 | |
| Chao1 | 1 709.26±868.06bc | 2 043.13±914.34bc | 2 674.50±1 066.06b | 8 847.05±1 696.64a | 837.88±332.8c | 108.4 | <0.01 |
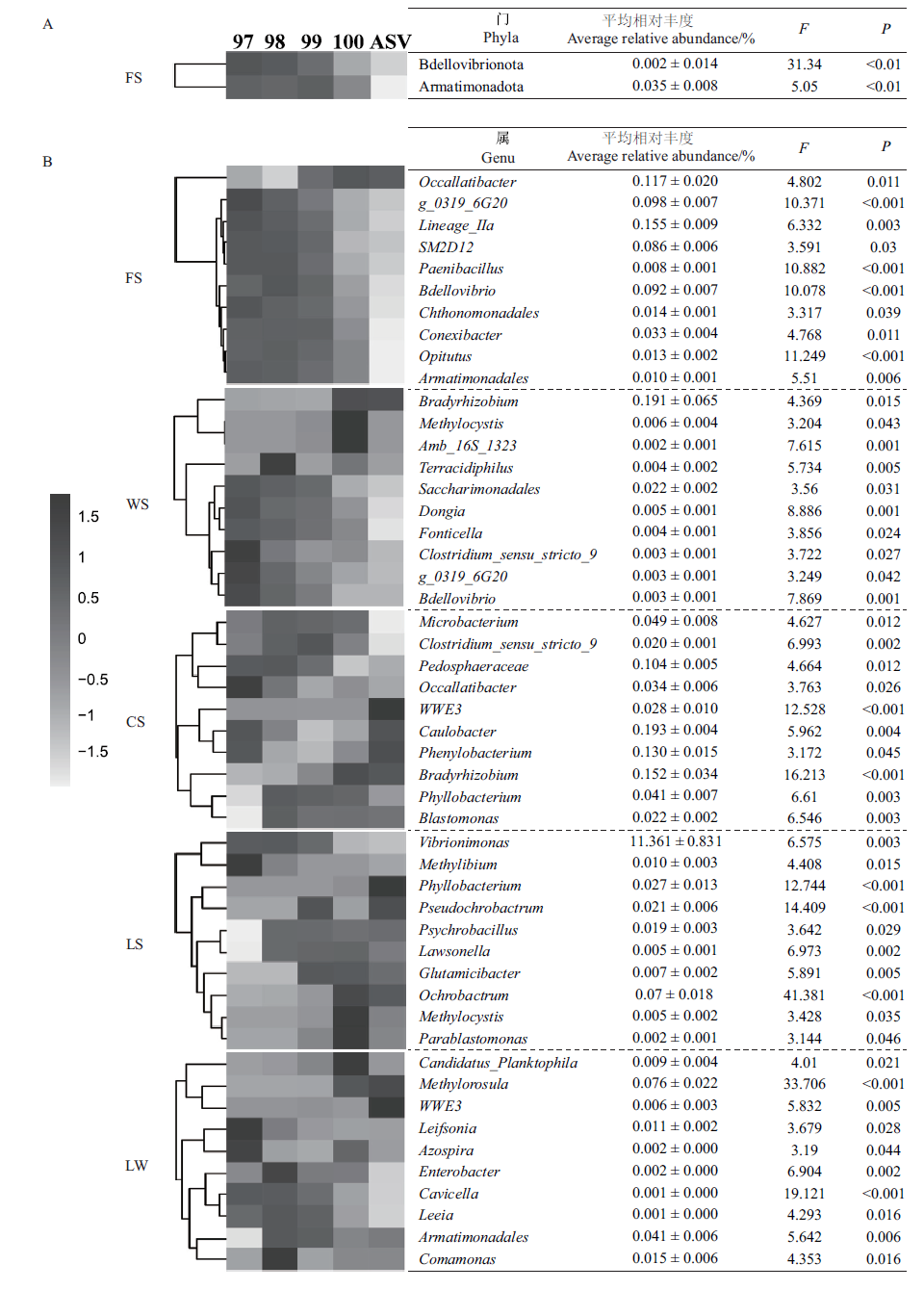
图1 最小分类单元划分方法对细菌群落门(A)和属(B)相对丰度的影响
Fig.1 Effects of the minimum taxonomy unit division method on the relative abundance of bacteria community phylum(A)and genus(B)
| 样地 Sampling site | 门总数 Number of phyla | 差异门 Differential phylum | 差异门丰度Abundance of differential phylum/% | 属总数 Number of genera | 差异属 Differential genus | 差异属丰度Abundance of differential genus/% |
|---|---|---|---|---|---|---|
| FS | 40 | 2 | 3.7 | 888 | 75 | 2.9 |
| WS | 52 | 0 | 0 | 854 | 30 | 0.35 |
| CS | 58 | 0 | 0 | 1 227 | 38 | 3.21 |
| LS | 54 | 0 | 0 | 1 304 | 15 | 14.9 |
| LW | 51 | 0 | 0 | 1 179 | 18 | 1.32 |
表4 最小分类单元划分方法对细菌群落门和属水平物种丰度的影响
Table 4 Effects of the minimum taxonomy unit division method on the abundances of bacterial community phylum and genus
| 样地 Sampling site | 门总数 Number of phyla | 差异门 Differential phylum | 差异门丰度Abundance of differential phylum/% | 属总数 Number of genera | 差异属 Differential genus | 差异属丰度Abundance of differential genus/% |
|---|---|---|---|---|---|---|
| FS | 40 | 2 | 3.7 | 888 | 75 | 2.9 |
| WS | 52 | 0 | 0 | 854 | 30 | 0.35 |
| CS | 58 | 0 | 0 | 1 227 | 38 | 3.21 |
| LS | 54 | 0 | 0 | 1 304 | 15 | 14.9 |
| LW | 51 | 0 | 0 | 1 179 | 18 | 1.32 |
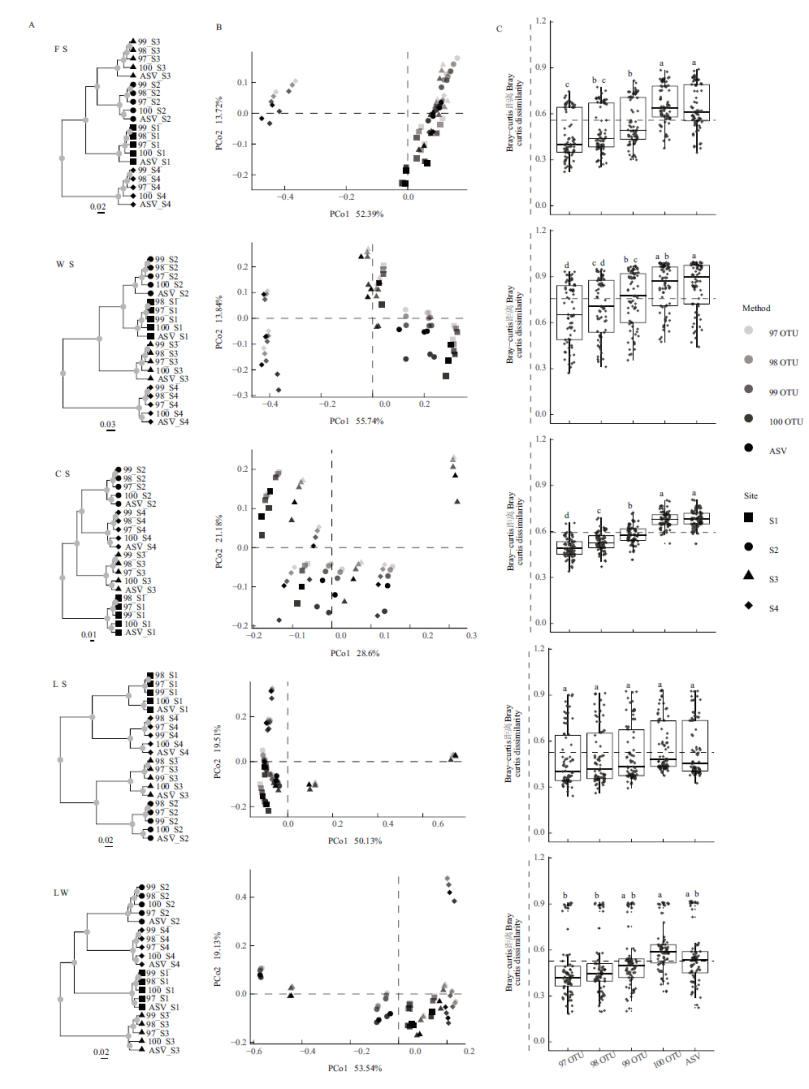
图2 基于细菌群落(属水平)bray-curtis距离的β多样性分析 A:聚类分析;B:主坐标分析(PCoA);C:不同方法的β多样性差异
Fig.2 Analysis of β diversity based on the bacterial community(genus level)bray-curtis dissimility A:Cluster analysis. B:Principal co-ordinates analysis(PCoA). C:Differences in beta-diversity among the division methods
| [1] |
Locey KJ, Lennon JT. Scaling laws predict global microbial diversity[J]. PNAS, 2016, 113(21):5970-5975.
doi: 10.1073/pnas.1521291113 URL |
| [2] |
高贵锋, 褚海燕. 微生物组学的技术和方法及其应用[J]. 植物生态学报, 2020, 44(4):395-408.
doi: 10.17521/cjpe.2019.0222 |
|
Gao GF, Chu HY. Techniques and methods of microbiomics and their applications[J]. Chin J Plant Ecol, 2020, 44(4):395-408.
doi: 10.17521/cjpe.2019.0222 URL |
|
| [3] |
Sanz JL, Köchling T. Molecular biology techniques used in wastewater treatment:an overview[J]. Process Biochem, 2007, 42(2):119-133.
doi: 10.1016/j.procbio.2006.10.003 URL |
| [4] | Caruso V, Song XB, Asquith M, et al. Performance of microbiome sequence inference methods in environments with varying biomass[J]. mSystems, 2019, 4(1):e00163-e00118. |
| [5] |
Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis[J]. BMC Bioinformatics, 2016, 17:135.
doi: 10.1186/s12859-016-0992-y pmid: 27000765 |
| [6] |
Loman NJ, Misra RV, Dallman TJ, et al. Performance comparison of benchtop high-throughput sequencing platforms[J]. Nat Biotechnol, 2012, 30(5):434-439.
doi: 10.1038/nbt.2198 URL |
| [7] |
Allali I, Arnold JW, Roach J, et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome[J]. BMC Microbiol, 2017, 17(1):194.
doi: 10.1186/s12866-017-1101-8 pmid: 28903732 |
| [8] |
Pedrós-Alió C. Marine microbial diversity:can it be determined?[J]. Trends Microbiol, 2006, 14(6):257-263.
pmid: 16679014 |
| [9] |
Lu HP, Yeh YC, Sastri AR, et al. Evaluating community-environment relationships along fine to broad taxonomic resolutions reveals evolutionary forces underlying community assembly[J]. Isme J, 2016, 10(12):2867-2878.
doi: 10.1038/ismej.2016.78 URL |
| [10] |
Capunitan DC, Johnson O, Terrill RS, et al. Evolutionary signal in the gut microbiomes of 74 bird species from Equatorial Guinea[J]. Mol Ecol, 2020, 29(4):829-847.
doi: 10.1111/mec.15354 pmid: 31943484 |
| [11] |
Edgar RC. UPARSE:highly accurate OTU sequences from microbial amplicon reads[J]. Nat Methods, 2013, 10(10):996-998.
doi: 10.1038/nmeth.2604 URL |
| [12] |
Gevers D, Cohan FM, Lawrence JG, et al. Re-evaluating prokaryotic species[J]. Nat Rev Microbiol, 2005, 3(9):733-739.
pmid: 16138101 |
| [13] | Erko S, Ebers J. Taxonomic parameters revisited:tarnished gold standards[J]. Microbiol Today, 2006, 33:152-155. |
| [14] |
Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs[J]. Bioinformatics, 2018, 34(14):2371-2375.
doi: 10.1093/bioinformatics/bty113 URL |
| [15] |
Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2:High-resolution sample inference from Illumina amplicon data[J]. Nat Methods, 2016, 13(7):581-583.
doi: 10.1038/NMETH.3869 |
| [16] |
Berg G, Rybakova D, et al. Microbiome definition re-visited:old concepts and new challenges[J]. Microbiome, 2020, 8(1):103.
doi: 10.1186/s40168-020-00875-0 URL |
| [17] |
Scarlett K, Denman S, Clark DR, et al. Relationships between nitrogen cycling microbial community abundance and composition reveal the indirect effect of soil pH on oak decline[J]. Isme J, 2021, 15(3):623-635.
doi: 10.1038/s41396-020-00801-0 pmid: 33067585 |
| [18] |
Joos L, Beirinckx S, Haegeman A, et al. Daring to be differential:metabarcoding analysis of soil and plant-related microbial communities using amplicon sequence variants and operational taxonomical units[J]. BMC Genom, 2020, 21(1):733.
doi: 10.1186/s12864-020-07126-4 URL |
| [19] | 刘亚军, 蔡润发, 李赟璟, 等. 湿地土壤微生物碳源代谢活性对不同水分条件的动态响应——以鄱阳湖为例[J]. 土壤, 2018, 50(4):705-711. |
| Liu YJ, Cai RF, Li YJ, et al. Functional response of wetland soil microbial carbon source metabolic activity to different water conditions—A case of lake Poyang[J]. Soils, 2018, 50(4):705-711. | |
| [20] | 何世耀. 鄱阳湖饶河入湖口水体和底泥微生物对氮、磷及重金属污染物输入的响应[D]. 南昌: 南昌大学, 2019. |
| He SY. The response of microbial in sediment and water of Poyang lake to the inputs of N, P and heavy metals pollutions in Raohe river[D]. Nanchang: Nanchang University, 2019. | |
| [21] |
Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2[J]. Nat Biotechnol, 2019, 37(8):852-857.
doi: 10.1038/s41587-019-0209-9 pmid: 31341288 |
| [22] | Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads[J]. EMBnet J, 2011, 17(1):10. |
| [23] | Rognes T, Flouri T, Nichols B, et al. VSEARCH:a versatile open source tool for metagenomics[J]. PeerJ, 2016, 4:e2584. |
| [24] | Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project:improved data processing and web-based tools[J]. Nucleic Acids Res, 2013, 41(database issue):D590-D596. |
| [25] |
Bokulich NA, Kaehler BD, Rideout JR, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin[J]. Microbiome, 2018, 6(1):90.
doi: 10.1186/s40168-018-0470-z URL |
| [26] |
Katoh K, Misawa K, Kuma K, et al. MAFFT:a novel method for rapid multiple sequence alignment based on fast Fourier transform[J]. Nucleic Acids Res, 2002, 30(14):3059-3066.
doi: 10.1093/nar/gkf436 URL |
| [27] | Price MN, Dehal PS, Arkin AP. FastTree 2——approximately maximum-likelihood trees for large alignments[J]. PLoS One, 2010, 5(3):e9490. |
| [28] | Oksanen J, Blanchet FG, Friendly M, et al. vegan:Community Ecology Package[J]. R package version 2. 5-7, 2020. |
| [29] |
Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models[J]. Biom J, 2008, 50(3):346-363.
doi: 10.1002/bimj.200810425 URL |
| [30] |
Wickham H. ggplot2[J]. WIREs Comp Stat, 2011, 3(2):180-185.
doi: 10.1002/wics.147 URL |
| [31] |
Shade A. Diversity is the question, not the answer[J]. Isme J, 2017, 11(1):1-6.
doi: 10.1038/ismej.2016.118 URL |
| [32] | Louca S, Mazel F, Doebeli M, et al. A census-based estimate of Earth’s bacterial and archaeal diversity[J]. PLoS Biol, 2019, 17(2):e3000106. |
| [33] | Li XC, Huo SL, Xi BD. Updating the resolution for 16S rRNA OTUs clustering reveals the cryptic cyanobacterial genus and species[J]. Ecol Indic, 2020, 117:106695. |
| [34] |
Kunin V, Engelbrektson A, et al. Wrinkles in the rare biosphere:pyrosequencing errors can lead to artificial inflation of diversity estimates[J]. Environ Microbiol, 2010, 12(1):118-123.
doi: 10.1111/j.1462-2920.2009.02051.x URL |
| [35] |
Washburne AD, Morton JT, Sanders J, et al. Methods for phylogenetic analysis of microbiome data[J]. Nat Microbiol, 2018, 3(6):652-661.
doi: 10.1038/s41564-018-0156-0 pmid: 29795540 |
| [36] |
Barnes CJ, Rasmussen L, et al. Comparing DADA2 and OTU clustering approaches in studying the bacterial communities of atopic dermatitis[J]. J Med Microbiol, 2020, 69(11):1293-1302.
doi: 10.1099/jmm.0.001256 URL |
| [37] |
Zhou Z, Wang C, Luo Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality[J]. Nat Commun, 2020, 11(1):3072.
doi: 10.1038/s41467-020-16881-7 URL |
| [38] |
Berlow M, Phillips JN, Derryberry EP. Effects of urbanization and landscape on gut microbiomes in white-crowned sparrows[J]. Microb Ecol, 2021, 81(1):253-266.
doi: 10.1007/s00248-020-01569-8 URL |
| [39] |
Cottrell MT, David KL. Contribution of major bacterial groups to bacterial biomass production(thymidine and leucine incorporation)in the Delaware estuary[J]. Limnol Oceanogr, 2003, 48(1):168-178.
doi: 10.4319/lo.2003.48.1.0168 URL |
| [40] | Dell’Anno F, Rastelli E, et al. Highly contaminated marine sediments can host rare bacterial taxa potentially useful for bioremediation[J]. Front Microbiol, 2021, 12:584850. |
| [41] |
Grady EN, MacDonald J, Liu L, et al. Current knowledge and perspectives of Paenibacillus:a review[J]. Microb Cell Fact, 2016, 15(1):203.
doi: 10.1186/s12934-016-0603-7 URL |
| [42] |
McSpadden Gardener BB. Ecology of Bacillus and Paenibacillus spp. in agricultural systems[J]. Phytopathology®, 2004, 94(11):1252-1258.
doi: 10.1094/PHYTO.2004.94.11.1252 URL |
| [43] |
Monciardini P, Cavaletti L, Schumann P, et al. Conexibacter woesei gen. nov., sp. nov., a novel representative of a deep evolutionary line of descent within the class Actinobacteria[J]. Int J Syst Evol Microbiol, 2003, 53(Pt 2):569-576.
doi: 10.1099/ijs.0.02400-0 URL |
| [44] | Dong L, Cheng R, Xiao L, et al. Diversity and composition of bacterial endophytes among plant parts of Panax notoginseng[J]. Chinese medicine, 2018, 13(1):1-9. |
| [45] |
Berestovskaya JJ, Kotsyurbenko OR, Tourova TP, et al. Methylorosula polaris gen. nov., sp. nov., an aerobic, facultatively methylotrophic psychrotolerant bacterium from tundra wetland soil[J]. Int J Syst Evol Microbiol, 2012, 62(Pt 3):638-646.
doi: 10.1099/ijs.0.007005-0 URL |
| [46] |
Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities[J]. Appl Environ Microbiol, 2007, 73(5):1576-1585.
doi: 10.1128/AEM.01996-06 URL |
| [47] |
Tromas N, Fortin N, Bedrani L, et al. Characterising and predicting cyanobacterial blooms in an 8-year amplicon sequencing time course[J]. Isme J, 2017, 11(8):1746-1763.
doi: 10.1038/ismej.2017.58 URL |
| [48] |
An JX, Liu C, Wang Q, et al. Soil bacterial community structure in Chinese wetlands[J]. Geoderma, 2019, 337:290-299.
doi: 10.1016/j.geoderma.2018.09.035 URL |
| [1] | 李琦, 杨晓蕾, 李晓林, 申友磊, 李建宏, 姚拓. 高寒草地燕麦根际解植酸磷促生菌鉴定及其优势菌假单胞菌属菌株功能特性[J]. 生物技术通报, 2023, 39(3): 243-253. |
| [2] | 孙海航, 官会林, 王旭, 王童, 李泓霖, 彭文洁, 刘柏桢, 樊芳玲. 生物炭对三七连作土壤性质及真菌群落的影响[J]. 生物技术通报, 2023, 39(2): 221-231. |
| [3] | 王子夜, 王志刚, 阎爱华. 不同树龄桑根际土壤原生生物群落组成多样性[J]. 生物技术通报, 2022, 38(8): 206-215. |
| [4] | 陈天赐, 武少兰, 杨国辉, 江丹霞, 江玉姬, 陈炳智. 无柄灵芝醇提物对小鼠睡眠及肠道菌群的影响[J]. 生物技术通报, 2022, 38(8): 225-232. |
| [5] | 赵林艳, 官会林, 向萍, 李泽诚, 柏雨龙, 宋洪川, 孙世中, 徐武美. 白及根腐病植株根际土壤微生物群落组成特征分析[J]. 生物技术通报, 2022, 38(2): 67-74. |
| [6] | 陈宇捷, 郑华宝, 周昕彦. 改良高通量测序技术揭示除藻剂对藻类群落的影响[J]. 生物技术通报, 2022, 38(11): 70-79. |
| [7] | 曹修凯, 王珊, 葛玲, 张卫博, 孙伟. 染色体外环形DNA研究进展及其在畜禽育种中的应用[J]. 生物技术通报, 2022, 38(1): 247-257. |
| [8] | 王琦, 武之绚, 陈钟玲, 吴白乙拉, 胡宗福, 牛化欣. 副干酪乳杆菌对青贮苜蓿有氧暴露品质和细菌多样性的影响[J]. 生物技术通报, 2021, 37(9): 77-85. |
| [9] | 毛婷, 牛永艳, 郑群, 杨涛, 穆永松, 祝英, 季彬, 王治业. 菌剂对苜蓿青贮发酵品质及微生物群落的影响[J]. 生物技术通报, 2021, 37(9): 86-94. |
| [10] | 唐蝶, 周倩. 植物基因组组装技术研究进展[J]. 生物技术通报, 2021, 37(6): 1-12. |
| [11] | 吕燕, 刘建利, 李靖宇, 候琳琳, 孙敏, 苟琪. 不同品种和产区宁夏枸杞根系AMF多样性[J]. 生物技术通报, 2021, 37(6): 36-48. |
| [12] | 朱斌, 甘晨晨, 王洪程. 球花石斛(Dendrobium thyrsiflorum)叶绿体基因组特征及亲缘关系解析[J]. 生物技术通报, 2021, 37(5): 38-47. |
| [13] | 张秫华, 方千, 贾红梅, 韩桂琪, 严铸云, 何冬梅. 川芎非根际、根际及根茎内生真菌群落差异分析[J]. 生物技术通报, 2021, 37(4): 56-69. |
| [14] | 郭艳萍, 张浩, 赵新钢, 罗海玲, 张英俊. DNA宏条形码技术在食草动物食性研究中的应用[J]. 生物技术通报, 2021, 37(3): 252-260. |
| [15] | 郑芳芳, 林俊生. 增殖诱导配体蛋白的核酸适配体筛选与特异性研究[J]. 生物技术通报, 2021, 37(10): 196-202. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||