








生物技术通报 ›› 2022, Vol. 38 ›› Issue (6): 93-102.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1058
聂立斌1,2( ), 易铃欣1,2, 邓妍1,2, 盛琦1,2, 吴晓玉1,2, 张斌1,2(
), 易铃欣1,2, 邓妍1,2, 盛琦1,2, 吴晓玉1,2, 张斌1,2( )
)
收稿日期:2021-08-18
出版日期:2022-06-26
发布日期:2022-07-11
作者简介:聂立斌,男,硕士研究生,研究方向:微生物代谢工程;E-mail: 基金资助:
NIE Li-bin1,2( ), YI Ling-xin1,2, DENG Yan1,2, SHENG Qi1,2, WU Xiao-yu1,2, ZHANG Bin1,2(
), YI Ling-xin1,2, DENG Yan1,2, SHENG Qi1,2, WU Xiao-yu1,2, ZHANG Bin1,2( )
)
Received:2021-08-18
Published:2022-06-26
Online:2022-07-11
摘要:
莽草酸是一种芳香族中间代谢产物,也是合成抗禽流感药物磷酸奥司他韦的前体。目前,国内外莽草酸的生产主要依靠成本较高,周期较长的植物提取法。微生物发酵法合成莽草酸具有生产成本低、周期短等优势成为研究的热点。为了构建产莽草酸的重组谷氨酸棒杆菌,此次研究从基因组水平上对谷氨酸棒杆菌体内的莽草酸代谢途径进行代谢工程改造。通过阻断莽草酸分解代谢途径、解除反馈抑制以及阻断竞争性代谢途径的策略,实现了莽草酸产量的大幅提升。结果显示,所构建的重组谷氨酸棒杆菌SKA06经72 h摇瓶发酵,莽草酸产量达到7.61 g/L,相较出发菌种提升了68倍。并且,基于染色体工程的遗传改造策略克服了引入质粒带来传代不稳定、需要添加抗生素等问题,可以为莽草酸工程菌种的选育提供重要参考。
聂立斌, 易铃欣, 邓妍, 盛琦, 吴晓玉, 张斌. 途径工程改造谷氨酸棒杆菌产莽草酸[J]. 生物技术通报, 2022, 38(6): 93-102.
NIE Li-bin, YI Ling-xin, DENG Yan, SHENG Qi, WU Xiao-yu, ZHANG Bin. Pathway Engineering Modification of Corynebacterium glutamicum for Shikimic Acid Production[J]. Biotechnology Bulletin, 2022, 38(6): 93-102.
| Strain | Relevant genotype | Reference |
|---|---|---|
| Escherichia coli DH5α | Cloning host | Lab stock |
| Corynebacterium. glutamicum CICC 20189 | Wild type,L-phenylalanine producing strain | Purchased from CICC |
| SKA01 | CICC 20189 with deletion of aroK | This study |
| SKA02 | CICC 20189 with deletion of aroK and a strong PNCgl0824 promoter insertion in the upstream region of aroG | This study |
| SKA03 | CICC 20189 with deletion of aroK and a strong Psod promoter insertion in the upstream region of aroG | This study |
| SKA04 | CICC 20189 with deletion of aroK,a strong PNCgl0824 promoter insertion in the upstream region of aroG,and deletion of qsuB | This study |
| SKA05 | CICC 20189 with deletion of aroK,a strong PNCgl0824 promoter insertion in the upstream region of aroG,deletion of qsuB,and insertion of SaroGE.coli at the △qsuB site. | This study |
| SKA06 | CICC 20189 with deletion of aroK,a strong PNCgl0824 promoter insertion in the upstream region of aroG,deletion of qsuB,and insertion of ParoGE.coli at the △qsuB site. | This study |
表1 本实验所用的菌种
Table 1 Corynebacterium glutamicum and Escherichia coli strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| Escherichia coli DH5α | Cloning host | Lab stock |
| Corynebacterium. glutamicum CICC 20189 | Wild type,L-phenylalanine producing strain | Purchased from CICC |
| SKA01 | CICC 20189 with deletion of aroK | This study |
| SKA02 | CICC 20189 with deletion of aroK and a strong PNCgl0824 promoter insertion in the upstream region of aroG | This study |
| SKA03 | CICC 20189 with deletion of aroK and a strong Psod promoter insertion in the upstream region of aroG | This study |
| SKA04 | CICC 20189 with deletion of aroK,a strong PNCgl0824 promoter insertion in the upstream region of aroG,and deletion of qsuB | This study |
| SKA05 | CICC 20189 with deletion of aroK,a strong PNCgl0824 promoter insertion in the upstream region of aroG,deletion of qsuB,and insertion of SaroGE.coli at the △qsuB site. | This study |
| SKA06 | CICC 20189 with deletion of aroK,a strong PNCgl0824 promoter insertion in the upstream region of aroG,deletion of qsuB,and insertion of ParoGE.coli at the △qsuB site. | This study |
| Plasmid | Characteristics | Reference |
|---|---|---|
| pK18mobsacB | Mobilizable vector,allows for selection of double crossover in C. glutamicum,KmR,sacB | Lab stock |
| pK18-△aroK | A derivative of pK18mobsacB,harboring △aroK fragment | This study |
| pK18-ParoG | A derivative of pK18mobsacB,harboring ParoG fragment | This study |
| pK18-SaroG | A derivative of pK18mobsacB,harboring SaroG fragment | This study |
| pK18-△qsuB | A derivative of pK18mobsacB,harboring △qsuB fragment | This study |
| pK18-△qsuB-SaroGE.coli | A derivative of pK18mobsacB,harboring △qsuB-SaroGE.coli fragment | This study |
| pK18-△qsuB-ParoGE.coli | A derivative of pK18mobsacB,harboring △qsuB-ParoGE.coli fragment | This study |
表2 本实验所用的质粒
Table 2 Plasmids used in this study
| Plasmid | Characteristics | Reference |
|---|---|---|
| pK18mobsacB | Mobilizable vector,allows for selection of double crossover in C. glutamicum,KmR,sacB | Lab stock |
| pK18-△aroK | A derivative of pK18mobsacB,harboring △aroK fragment | This study |
| pK18-ParoG | A derivative of pK18mobsacB,harboring ParoG fragment | This study |
| pK18-SaroG | A derivative of pK18mobsacB,harboring SaroG fragment | This study |
| pK18-△qsuB | A derivative of pK18mobsacB,harboring △qsuB fragment | This study |
| pK18-△qsuB-SaroGE.coli | A derivative of pK18mobsacB,harboring △qsuB-SaroGE.coli fragment | This study |
| pK18-△qsuB-ParoGE.coli | A derivative of pK18mobsacB,harboring △qsuB-ParoGE.coli fragment | This study |
| Primer name | Sequence(5'-3') | Size/bp |
|---|---|---|
| aroK-up-F | aacgacggccagtgccaagctCAATCGATGATTCCCCAGTTC | 42 |
| aroK -up-R | GAAAAGCTTGGGAAACGTCTAGAGCGTCGGAGTCGACGAGTTCAGTG | 47 |
| aroK -down-F | GCTCTAGACGTTTCCCAAGCTTTTCAAGCACTGAAATCCTCCGGAGT | 47 |
| aroK -down-R | cggtacccggggatcctctagCGGTGTCAATGAATACTGCGTC | 43 |
| aroK -check-F | AAAAGCCTGTGGCGCCGTGTT | 21 |
| aroG-up-F | aacgacggccagtgccaagctCTTGTGAGCGCTTCTTTGATC | 42 |
| aroG-up-R(sod) | CAAGCCCGGAATAATTGGCAATGGGATGGGGTGAATTTAGG | 41 |
| aroG-up-R(P) | CTATCCGTATTAGTGGCACAGTTATGGGATGGGGTGAATTTAGG | 44 |
| sod-F | TGCCAATTATTCCGGGCTTG | 20 |
| sod-R | TCCGCACCGAGCATATACATCTT | 23 |
| PNCgl0824-F | AACTGTGCCACTAATACGGATAG | 23 |
| PNCgl0824-R | CAATTTCGCCTGCTTCCGATT | 21 |
| aroG-down-F(sod) | AAGATGTATATGCTCGGTGCGGATGCATAGCCCTGAAAGGCAAG | 44 |
| aroG-down-F(P) | AATCGGAAGCAGGCGAAATTGTGCATAGCCCTGAAAGGCAAG | 42 |
| aroG-down-R | cggtacccggggatcctctagTCGTCGGAGGTTCCGAAGAAG | 42 |
| qsuB-up-F | aacgacggccagtgccaagctGCTCGCGTTGCTATTGCTG | 40 |
| qsuB-up-R | GAAAAGCTTGGGAAACGTCTAGAGCGAAACCACCAAGTCCTGCTCG | 46 |
| qsuB-down-F | GCTCTAGACGTTTCCCAAGCTTTTCATCTGTTCCAGCCGTTTCGAG | 46 |
| qsuB-down-R | cggtacccggggatcctctagCACCCAAGCGGAAACCCAATT | 42 |
| qsuB-check-F | GGCTCAGGATTTGGGATTAAC | 21 |
| Sod-F(qsuB) | ACTTGGTGGTTTCGCTCTAGATGCCAATTATTCCGGGCTTG | 41 |
| Sod-R(aroGE. coli) | AAATCGTCGTTCTGATAATTCATTCCGCACCGAGCATATACATCTT | 46 |
| P-F(qsuB) | ACTTGGTGGTTTCGCTCTAGAAACTGTGCCACTAATACGGATAG | 44 |
| P-R(aroGE. coli) | AAATCGTCGTTCTGATAATTCATCAATTTCGCCTGCTTCCGATT | 44 |
| aroGE. coli-F1 | ATGAATTATCAGAACGACGATTT | 23 |
| aroGE. coli-R1 | CGACCGGACAAAAAAGCCCTGATGCCAGTTCG | 32 |
| aroGE. coli-F2 | GCATCAGGGCTTTTTTGTCCGGTCGGCTTCAAAAATG | 37 |
| aroGE. coli-R2 | ACGGCTGGAACAGATGAAAAGCTTTTACCCGCGACGCGCTTTTACT | 46 |
表3 本实验所用的引物
Table 3 Primers used in this study
| Primer name | Sequence(5'-3') | Size/bp |
|---|---|---|
| aroK-up-F | aacgacggccagtgccaagctCAATCGATGATTCCCCAGTTC | 42 |
| aroK -up-R | GAAAAGCTTGGGAAACGTCTAGAGCGTCGGAGTCGACGAGTTCAGTG | 47 |
| aroK -down-F | GCTCTAGACGTTTCCCAAGCTTTTCAAGCACTGAAATCCTCCGGAGT | 47 |
| aroK -down-R | cggtacccggggatcctctagCGGTGTCAATGAATACTGCGTC | 43 |
| aroK -check-F | AAAAGCCTGTGGCGCCGTGTT | 21 |
| aroG-up-F | aacgacggccagtgccaagctCTTGTGAGCGCTTCTTTGATC | 42 |
| aroG-up-R(sod) | CAAGCCCGGAATAATTGGCAATGGGATGGGGTGAATTTAGG | 41 |
| aroG-up-R(P) | CTATCCGTATTAGTGGCACAGTTATGGGATGGGGTGAATTTAGG | 44 |
| sod-F | TGCCAATTATTCCGGGCTTG | 20 |
| sod-R | TCCGCACCGAGCATATACATCTT | 23 |
| PNCgl0824-F | AACTGTGCCACTAATACGGATAG | 23 |
| PNCgl0824-R | CAATTTCGCCTGCTTCCGATT | 21 |
| aroG-down-F(sod) | AAGATGTATATGCTCGGTGCGGATGCATAGCCCTGAAAGGCAAG | 44 |
| aroG-down-F(P) | AATCGGAAGCAGGCGAAATTGTGCATAGCCCTGAAAGGCAAG | 42 |
| aroG-down-R | cggtacccggggatcctctagTCGTCGGAGGTTCCGAAGAAG | 42 |
| qsuB-up-F | aacgacggccagtgccaagctGCTCGCGTTGCTATTGCTG | 40 |
| qsuB-up-R | GAAAAGCTTGGGAAACGTCTAGAGCGAAACCACCAAGTCCTGCTCG | 46 |
| qsuB-down-F | GCTCTAGACGTTTCCCAAGCTTTTCATCTGTTCCAGCCGTTTCGAG | 46 |
| qsuB-down-R | cggtacccggggatcctctagCACCCAAGCGGAAACCCAATT | 42 |
| qsuB-check-F | GGCTCAGGATTTGGGATTAAC | 21 |
| Sod-F(qsuB) | ACTTGGTGGTTTCGCTCTAGATGCCAATTATTCCGGGCTTG | 41 |
| Sod-R(aroGE. coli) | AAATCGTCGTTCTGATAATTCATTCCGCACCGAGCATATACATCTT | 46 |
| P-F(qsuB) | ACTTGGTGGTTTCGCTCTAGAAACTGTGCCACTAATACGGATAG | 44 |
| P-R(aroGE. coli) | AAATCGTCGTTCTGATAATTCATCAATTTCGCCTGCTTCCGATT | 44 |
| aroGE. coli-F1 | ATGAATTATCAGAACGACGATTT | 23 |
| aroGE. coli-R1 | CGACCGGACAAAAAAGCCCTGATGCCAGTTCG | 32 |
| aroGE. coli-F2 | GCATCAGGGCTTTTTTGTCCGGTCGGCTTCAAAAATG | 37 |
| aroGE. coli-R2 | ACGGCTGGAACAGATGAAAAGCTTTTACCCGCGACGCGCTTTTACT | 46 |
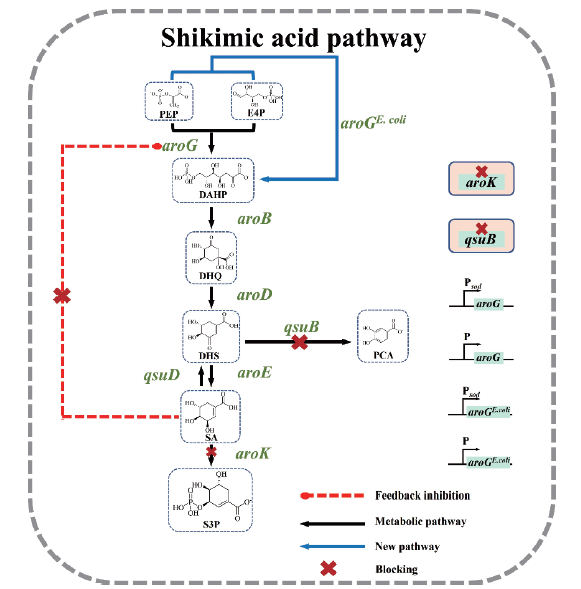
图1 谷氨酸棒杆菌的莽草酸代谢途径
Fig.1 Pathway of shikimic acid in C. glutamicum PEP:phosphoenolpyruvate;E4P:erythrose-4-phosphate;DAHP:3-deoxy-D-arabinoheptulosonate-7-phosphate;DHQ:3-dehydro-quinate;DHS:3-dehydroshikimate;PCA:protocatechuate;S3P:shikimate-3-phosphate SA,shikimic acid;aroG:encoding DAHP synthetase;aroGE. coli:encoding DAHP synthetase from E. coli;aroB:encoding DHQ synthase;aroD:encoding DHQ dehydratase;aroE:encoding shikimate dehydrogenase;aroK:encoding shikimate kinase;qsuB:encoding DHS dehydratase;qsuD:encoding QA/ shikimate dehydrogenase
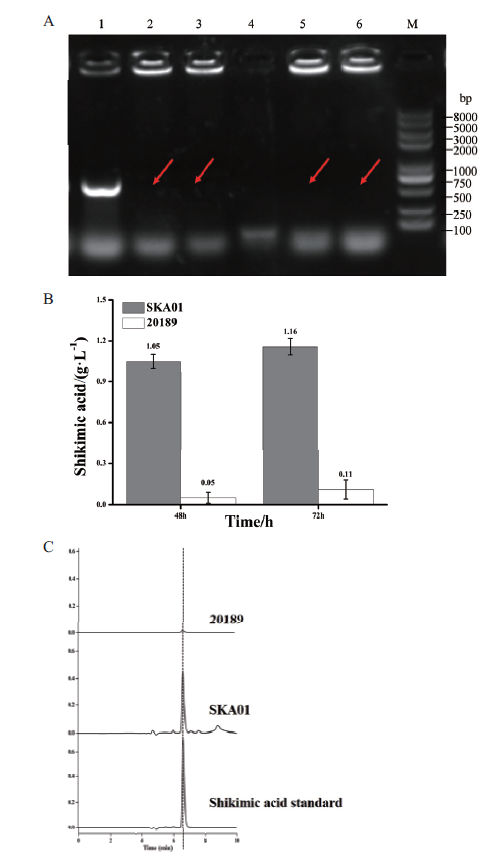
图2 aroK基因缺失株的构建及发酵性能评价 A:PCR鉴定aroK阳性基因敲除株;M:DNA marker;1:利用引物aroK-check-F和aroK-down-R扩增谷氨酸棒杆菌CICC 20189;4:阴性对照;2,3,5,6:利用引物aroK-check-F和aroK-down-R扩增谷氨酸棒杆菌SKA01。B:谷氨酸棒杆菌CICC 20189和SKA01发酵48和72 h的莽草酸产量。C:不同菌种产莽草酸的HPLC色谱图及标准品对照
Fig.2 Construction and fermentation evaluation of aroK-deleted strain A:PCR identification of positive strain with aroK deleted. M:DNA marker;1:amplifying C. glutamicum CICC 20189 using aroK-check-F and aroK-down-R as primers. 4:Negative control. 2,3,5,6:amplifying C. glutamicum SKA01 using aroK-check-F and aroK-down-R as primers. B:Shikimic acid production at fermented 48 h and 72 h of strain C. glutamicum CICC 20189 and SKA01.C:HPLC analysis of shikimic acid produced by different strains in contrast with standard sample
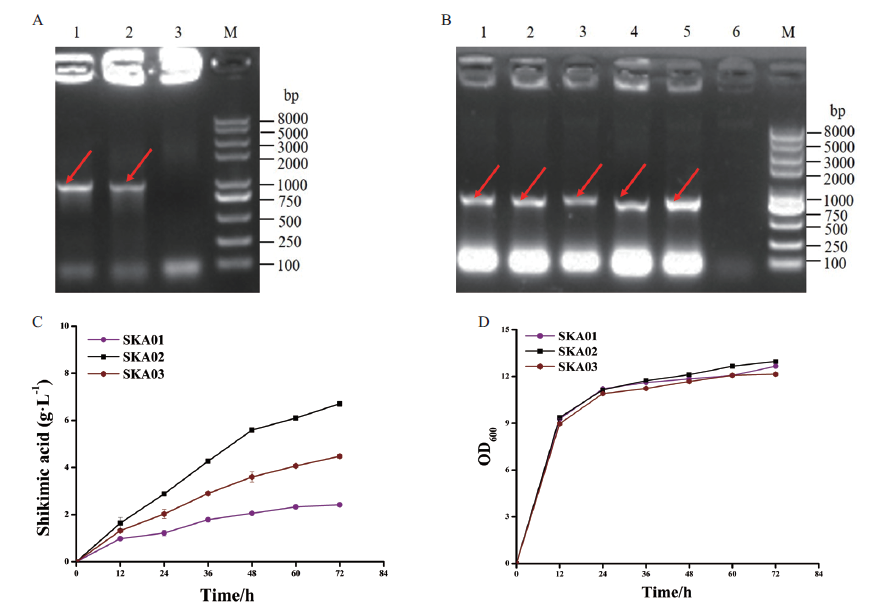
图3 重组谷氨酸棒杆菌SKA02和SKA03的PCR鉴定及摇瓶发酵性能评价 A:PCR鉴定重组菌SKA02;M:DNA marker;1,2:利用引物PNCgl0824-F和aroG-down-R扩增谷氨酸棒杆菌SKA02;3:利用引物PNCgl0824-F和aroG-down-R扩增谷氨酸棒杆菌SKA01;B:PCR鉴定重组菌SKA03;M:DNA marker;1-5:利用引物 sod-F和aroG-down-R扩增谷氨酸棒杆菌SKA03;6:利用引物sod-F和aroG-down-R扩增谷氨酸棒杆菌SKA01;C:谷氨酸棒杆菌SKA02和SKA03的莽草酸发酵曲线;D:谷氨酸棒杆菌SKA02和SKA03的生长曲线
Fig.3 PCR identification and shake flask fermentation evaluation of recombinant strain C. glutamicum SKA02 and SKA03 A:PCR identification of recombinant strain SKA02. M:DNA marker;1,2:amplifying C. glutamicum SKA02 using PNCgl0824-F and aroG-down-R as primers;3:amplifying C. glutamicum SKA01 using PNCgl0824-F and aroG-down-R as primers. B:PCR identification of recombinant strain SKA03. M:DNA marker;1-5:amplifying C. glutamicum SKA03 using sod-F and aroG-down-R;6:amplifying C. glutamicum SKA01 using sod-F and aroG-down-R. C:Shikimic acid production curves of C. glutamicum strain SKA02 and SKA03. D:Cell growth curves of C. glutamicum strain SKA02 and SKA03
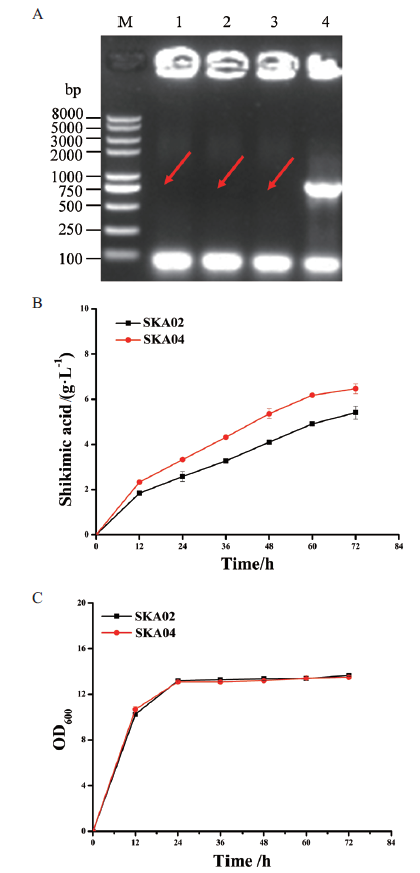
图4 重组谷氨酸棒杆菌SKA04的构建及发酵性能评价 A:PCR鉴定重组菌SKA04;M:DNA marker;1-3:利用引物qsuB-check-F和qsuB-down-R扩增谷氨酸棒杆菌SKA04;4:利用引物qsuB-check-F和qsuB-down-R扩增谷氨酸棒杆菌SKA02;B:谷氨酸棒杆菌SKA02和SKA04的莽草酸发酵曲线;C:谷氨酸棒杆菌SKA02和SKA04的生长曲线
Fig.4 Construction and fermentation evaluation of recom-binant C. glutamicum strain SKA04 A:PCR identification of recombinant SKA04. M:DNA marker;1-3:amplifying C. glutamicum SKA04 using qsuB-check-F and qsuB-down-R as primers;4:amplifying C. glutamicum SKA02 using qsuB-check-F and qsuB-down-R as primers. B:Shikimic acid production curves of C. glutamicum strain SKA02 and SKA04. C:Cell growth curves of C. glutamicum strain SKA02 and SKA04

图5 重组谷氨酸棒杆菌SKA04、SKA05和SKA06的构建及摇瓶发酵性能评价 A:PCR鉴定重组菌SKA05;M:DNA marker;1:利用引物PNCgl0824-F和aroGE. coli-R1扩增谷氨酸棒杆菌SKA04;2-8:利用引物PNCgl0824-F和aroGE. coli-R1扩增谷氨酸棒杆菌SKA05。B:PCR鉴定重组菌SKA06;M:DNA marker;1:利用引物 sod-F和aroGE. coli-R1扩增谷氨酸棒杆菌SKA06;2-8:利用引物sod-F和aroGE. coli-R1扩增谷氨酸棒杆菌SKA01。C:谷氨酸棒杆菌SKA04、SKA05和SKA06的莽草酸发酵曲线。D:谷氨酸棒杆菌SKA04、SKA05和SKA06的生长曲线
Fig. 5 Construction and shake flask fermentation evaluation of recombinant C. glutamicum strain SKA04,SKA05,and SKA06 A:PCR identification of SKA05. M:DNA marker;1:amplifying C. glutamicum SKA04 using PNCgl0824-F and aroGE. coli-R1 as primers;2-8:amplifying C. glutamicum SKA05 using PNCgl0824-F and aroGE. coli-R1 as primers. B:PCR identification of SKA06. M:DNA marker;1,2:amplifying C. glutamicum SKA06 using sod-F and aroGE. coli-R1 as primers;3:amplifying C. glutamicum SKA04 using sod-F and aroGE. coli-R1 as primers. C:Shikimic acid production curves of C. glutamicum strain SKA04,SKA05,and SKA06. D:Cell growth curves of C. glutamicum strain SKA04,SKA05,and SKA06
| [1] | 江晶洁, 刘涛, 林双君. 基于莽草酸途径微生物合成芳香族化合物及其衍生物的研究进展[J]. 生命科学, 2019, 31(5):430-448. |
| Jiang JJ, Liu T, Lin SJ. Research progress on the biosynthesis of aromatic compounds by microorganisms[J]. Chin Bull Life Sci, 2019, 31(5):430-448. | |
| [2] |
Gong J, Xu W. Different synthetic strategies of oseltamivir phosphate:a potent influenza neuraminidase inhibitor[J]. Curr Med Chem, 2008, 15(30):3145-3159.
doi: 10.2174/092986708786848497 URL |
| [3] |
Jiang M, Zhang H. Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli[J]. Curr Opin Biotechnol, 2016, 42:1-6.
doi: 10.1016/j.copbio.2016.01.016 URL |
| [4] | Adelfo Escalante A, Carmona SB, Diaz Quiroz DC, et al. Current perspectives on applications of shikimic and aminoshikimic acids in pharmaceutical chemistry[J]. Res Rep Med Chem, 2014, 4:35-46. |
| [5] |
Gibson JM, Thomas PS, Thomas JD, et al. Benzene-free synthesis of phenol[J]. Angew Chem Int Ed, 2001, 40(10):1945-1948.
pmid: 11385681 |
| [6] |
Candeias NR, Assoah B, Simeonov SP. Production and synthetic modifications of shikimic acid[J]. Chem Rev, 2018, 118(20):10458-10550.
doi: 10.1021/acs.chemrev.8b00350 pmid: 30350584 |
| [7] |
Sui R. Separation of shikimic acid from pine needles[J]. Chem Eng Technol, 2008, 31(3):469-473.
doi: 10.1002/ceat.200700413 URL |
| [8] | 何邵平, 禹琪芳, 段杰林, 等. 莽草酸的制备及其生物学功能研究进展[J]. 饲料博览, 2014(10):32-36. |
| He SP, Yu QF, Duan JL, et al. Advance on preparation and biological function of shikimic acid[J]. Feed Rev, 2014(10):32-36. | |
| [9] |
She DJ, Singh G. Chemical synthesis of shikimic acid and its analogues[J]. Tetrahedron, 1998, 54(19):4697-4753.
doi: 10.1016/S0040-4020(98)00016-7 URL |
| [10] |
Ghosh S, Chisti Y, Banerjee UC. Production of shikimic acid[J]. Biotechnol Adv, 2012, 30(6):1425-1431.
doi: 10.1016/j.biotechadv.2012.03.001 URL |
| [11] |
Rawat G, Tripathi P, Saxena RK. Expanding horizons of shikimic acid[J]. Appl Microbiol Biotechnol, 2013, 97(10):4277-4287.
doi: 10.1007/s00253-013-4840-y URL |
| [12] | McCrindle R, Overton KH, Raphael RA. 315. A stereospecific total synthesis ofD-(-)-shikimic acid[J]. J Chem Soc, 1960:1560-1565. |
| [13] |
Box JM, Harwood LM, Humphreys JL, et al. Dehydration of quinate derivatives:synthesis of a difluoromethylene homologue of shikimic acid[J]. Synlett, 2002, 2002(2):358-360.
doi: 10.1055/s-2002-19772 URL |
| [14] |
Zhang B, Liu ZQ, Liu C, et al. Application of CRISPRi in Corynebacterium glutamicum for shikimic acid production[J]. Biotechnol Lett, 2016, 38(12):2153-2161.
pmid: 27623797 |
| [15] |
Cui YY, Ling C, Zhang YY, et al. Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering[J]. Microb Cell Fact, 2014, 13:21.
doi: 10.1186/1475-2859-13-21 URL |
| [16] |
Cortés-Tolalpa L, Gutiérrez-Ríos RM, Martínez LM, et al. Global transcriptomic analysis of an engineered Escherichia coli strain lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system during shikimic acid production in rich culture medium[J]. Microb Cell Fact, 2014, 13(1):28.
doi: 10.1186/1475-2859-13-28 pmid: 24559297 |
| [17] |
Chen K, Dou J, Tang S, et al. Deletion of the aroK gene is essential for high shikimic acid accumulation through the shikimate pathway in E. coli[J]. Bioresour Technol, 2012, 119:141-147.
doi: 10.1016/j.biortech.2012.05.100 URL |
| [18] |
Chen X, Li M, Zhou L, et al. Metabolic engineering of Escherichia coli for improving shikimate synthesis from glucose[J]. Bioresour Technol, 2014, 166:64-71.
doi: 10.1016/j.biortech.2014.05.035 URL |
| [19] |
Liu DF, Ai GM, Zheng QX, et al. Metabolic flux responses to genetic modification for shikimic acid production by Bacillus subtilis strains[J]. Microb Cell Fact, 2014, 13(1):40.
doi: 10.1186/1475-2859-13-40 URL |
| [20] | Licona-Cassani C, Lara AR, Cabrera-Valladares N, et al. Inactivation of pyruvate kinase or the phosphoenolpyruvate:sugar phosphotransferase system increases shikimic and dehydroshikimic acid yields from glucose in Bacillus subtilis[J]. J Mol Microbiol Biotechnol, 2014, 24(1):37-45. |
| [21] |
Rawat G, Tripathi P, Yadav S, et al. An interactive study of influential parameters for shikimic acid production using statistical approach, scale up and its inhibitory action on different lipases[J]. Bioresour Technol, 2013, 144:675-679.
doi: 10.1016/j.biortech.2013.06.113 URL |
| [22] |
Tripathi P, Rawat G, Yadav S, et al. Fermentative production of shikimic acid:a paradigm shift of production concept from plant route to microbial route[J]. Bioprocess Biosyst Eng, 2013, 36(11):1665-1673.
doi: 10.1007/s00449-013-0940-4 URL |
| [23] |
Tripathi P, Rawat G, Yadav S, et al. Shikimic acid, a base compound for the formulation of swine/avian flu drug:statistical optimization, fed-batch and scale up studies along with its application as an antibacterial agent[J]. Antonie Van Leeuwenhoek, 2015, 107(2):419-431.
doi: 10.1007/s10482-014-0340-z pmid: 25563634 |
| [24] |
Kogure T, Kubota T, Suda M, et al. Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction[J]. Metab Eng, 2016, 38:204-216.
doi: 10.1016/j.ymben.2016.08.005 URL |
| [25] |
Zhang B, Zhou N, Liu YM, et al. Ribosome binding site libraries and pathway modules for shikimic acid synthesis with Corynebacterium glutamicum[J]. Microb Cell Fact, 2015, 14:71.
doi: 10.1186/s12934-015-0254-0 pmid: 25981633 |
| [26] |
Liu C, Zhang B, Liu YM, et al. New intracellular shikimic acid biosensor for monitoring shikimate synthesis in Corynebacterium glutamicum[J]. ACS Synth Biol, 2018, 7(2):591-601.
doi: 10.1021/acssynbio.7b00339 URL |
| [27] |
Zhang B, Jiang CY, Liu YM, et al. Engineering of a hybrid route to enhance shikimic acid production in Corynebacterium glutamicum[J]. Biotechnol Lett, 2015, 37(9):1861-1868.
doi: 10.1007/s10529-015-1852-y pmid: 25967037 |
| [28] |
Rawat G, Tripathi P, Jahan F, et al. A natural isolate producing shikimic acid:isolation, identification, and culture condition optimization[J]. Appl Biochem Biotechnol, 2013, 169(8):2290-2302.
doi: 10.1007/s12010-013-0150-1 URL |
| [29] |
Ghosh S, Banerjee UC. Generation of aroE overexpression mutant of Bacillus megaterium for the production of shikimic acid[J]. Microb Cell Fact, 2015, 14:69.
doi: 10.1186/s12934-015-0251-3 URL |
| [30] |
Wu XY, Guo XY, Zhang B, et al. Recent advances of L-ornithine biosynthesis in metabolically engineered Corynebacterium glutamicum[J]. Front Bioeng Biotechnol, 2019, 7:440.
doi: 10.3389/fbioe.2019.00440 URL |
| [31] |
Schäfer A, Tauch A, Jäger W, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19:selection of defined deletions in the chromosome of Corynebacterium glutamicum[J]. Gene, 1994, 145(1):69-73.
doi: 10.1016/0378-1119(94)90324-7 pmid: 8045426 |
| [32] |
Park SH, Kim HU, Kim TY, et al. Metabolic engineering of Corynebacterium glutamicum for L-arginine production[J]. Nat Commun, 2014, 5:4618.
doi: 10.1038/ncomms5618 URL |
| [33] | Man Z, Xu M, Rao Z, et al. Systems pathway engineering of Corynebacterium crenatum for improved L-arginine production[J]. Sci Rep, 2016, 6:28629. |
| [34] |
Xu M, Li J, Shu Q, et al. Enhancement of L-arginine production by increasing ammonium uptake in an AmtR-deficient Corynebacterium crenatum mutant[J]. J Ind Microbiol Biotechnol, 2019, 46(8):1155-1166.
doi: 10.1007/s10295-019-02204-3 URL |
| [35] |
Zhang B, Gao G, Chu XH, et al. Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of l-ornithine driven by glucose and xylose[J]. Bioresour Technol, 2019, 284:204-213.
doi: 10.1016/j.biortech.2019.03.122 URL |
| [36] |
Zhang J, Qian F, Dong F, et al. De novo engineering of Corynebacterium glutamicum for l-proline production[J]. ACS Synth Biol, 2020, 9(7):1897-1906.
doi: 10.1021/acssynbio.0c00249 pmid: 32627539 |
| [1] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [2] | 刘佳慧, 刘叶, 花尔并, 王猛. 谷氨酸棒杆菌中胞嘧啶碱基编辑工具的PAM拓展[J]. 生物技术通报, 2023, 39(9): 49-57. |
| [3] | 黄华媚, 白立宽, 刘叶, 李俊维, 王猛, 花尔并. BE3型胞嘧啶碱基编辑器在谷氨酸棒杆菌中的开发用[J]. 生物技术通报, 2020, 36(3): 95-101. |
| [4] | 聂志华, 朱蕾蕾. 生物素对发酵过程中MscCG外排L-谷氨酸的影响[J]. 生物技术通报, 2020, 36(10): 150-155. |
| [5] | 徐德雨, 郑小梅, 赵晶, 郑平, 赵树欣. 谷氨酸棒杆菌天冬氨酸激酶G359D突变解除赖氨酸与苏氨酸协同抑制的研究[J]. 生物技术通报, 2017, 33(11): 143-152. |
| [6] | 徐杰, 蒋世云, 傅凤鸣, 耿鹏飞, 黄凯. EPSP合酶的研究进展[J]. 生物技术通报, 2014, 0(6): 40-50. |
| [7] | 石增秀 崔文璟 周丽 周哲敏. 谷氨酸棒杆菌L-天冬氨酸α-脱羧酶基因的克隆及重组酶性质研究[J]. 生物技术通报, 2013, 0(4): 110-115. |
| [8] | 罗玉常;窦文芳;张晓梅;史劲松;许正宏;. 谷氨酸棒杆菌ilvE基因的敲除对相关氨基酸合成的影响[J]. , 2012, 0(11): 185-191. |
| [9] | 黄云雁;黎明;刘萌;孙昕;周丽颖;路福平;. 赖氨酸-尸胺反向转运蛋白cadB基因谷氨酸棒杆菌表达载体的构建及转化[J]. , 2012, 0(08): 94-100. |
| [10] | 伍展红;郑穗平;. 谷氨酸棒杆菌ldh基因的敲除[J]. , 2012, 0(02): 107-111. |
| [11] | 李智涛;伍展红;郑穗平;. 谷氨酸棒杆菌aroⅡ、trpEGD基因的过量表达及其对色氨酸合成的影响[J]. , 2011, 0(05): 151-156. |
| [12] | 吕扬勇;伍展红;郑穗平;. 谷氨酸棒杆菌metX、dapA基因敲除对苏氨酸合成的影响[J]. , 2011, 0(01): 158-164. |
| [13] | 汪华;崔志峰;. 莽草酸生物合成途径的调控[J]. , 2009, 0(03): 50-53. |
| [14] | 林琳;窦文芳;张晓梅;许泓瑜;许正宏;王正祥;. 直接利用糖质原料产L-丝氨酸谷氨酸棒杆菌glyA基因序列分析[J]. , 2008, 0(05): 176-180. |
| [15] | 苏建亚;沈晋良. 阿维菌素的生物合成与途径工程[J]. , 2003, 0(06): 18-23. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||