








生物技术通报 ›› 2022, Vol. 38 ›› Issue (11): 202-209.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1604
武林辉( ), 耿必苗, 王艳杰, 周国伟, 孙庆业, 赵琼(
), 耿必苗, 王艳杰, 周国伟, 孙庆业, 赵琼( )
)
收稿日期:2021-12-28
出版日期:2022-11-26
发布日期:2022-12-01
作者简介:武林辉,男,硕士研究生,研究方向:森林土壤磷循环;E-mail:基金资助:
WU Lin-hui( ), GENG Bi-miao, WANG Yan-jie, ZHOU Guo-wei, SUN Qing-ye, ZHAO Qiong(
), GENG Bi-miao, WANG Yan-jie, ZHOU Guo-wei, SUN Qing-ye, ZHAO Qiong( )
)
Received:2021-12-28
Published:2022-11-26
Online:2022-12-01
摘要:
为研究氮添加影响森林土壤有机磷矿化的微生物调控机制,分析了10年的野外氮添加(100 kg N ha-2year-1)对沙地樟子松人工林土壤微生物中编码酸性磷酸单酯酶、碱性磷酸单酯酶和植酸酶的功能基因(phoC、phoD和appA)丰度及相关酶活性和土壤理化性质的影响。结果表明,氮添加使樟子松人工林土壤中酸性和碱性磷酸单酯酶活性分别下降了18.09%和55.29%,植酸酶活性下降了41.88%。氮添加使土壤微生物中各基因拷贝数分别下降40.97%(16S-rRNA)、78.38%(phoD)、67.92%(phoC)、74.37%(appA)。各基因拷贝数占总细菌基因拷贝数的比例显著下降了61%(phoD)、44%(phoC)、55%(appA)。土壤微生物量碳、微生物量磷含量与酸性磷酸单脂酶、碱性磷酸单脂酶、植酸酶活性及16S rRNA、phoD、phoC、appA基因丰度显著正相关。土壤铵态氮含量与酸性磷酸单脂酶、碱性磷酸单脂酶活性及16S rRNA、phoC、appA基因丰度显著负相关。酸性磷酸单酯酶活性与其基因丰度显著正相关,其他两种酶活性与其基因丰度无显著相关性。以上结果表明,氮添加不仅降低了土壤解磷微生物丰度,还降低了解磷细菌在总细菌中的比例,从而大大降低了磷酸酶活性,抑制了有机磷的矿化。
武林辉, 耿必苗, 王艳杰, 周国伟, 孙庆业, 赵琼. 氮添加对樟子松人工林土壤细菌磷酸酶编码基因丰度的影响[J]. 生物技术通报, 2022, 38(11): 202-209.
WU Lin-hui, GENG Bi-miao, WANG Yan-jie, ZHOU Guo-wei, SUN Qing-ye, ZHAO Qiong. Effects of Nitrogen Addition on the Abundance of Bacterial Phosphatase Encoding Genes in the Soil of Pinus sylvestris var. mongolica Plantation[J]. Biotechnology Bulletin, 2022, 38(11): 202-209.
| 引物Primer | 引物名称Primer name | 引物长度Primer length/bp | 引物序列Primer sequence(5'-3') | 参考文献Reference |
|---|---|---|---|---|
| phoD | ALPS-F730 | 370 | CAGTGGGACGA CCACGAGGT | [ |
| ALPS-1101 | GAGGCCGATCG GCATGTCG | |||
| phoC | phoC-A-F1 | 155 | CGGCTCCTATCCGTCCGG | [ |
| phoC-A-R1 | CAACATCGCTTTGCCAGTG | |||
| appA | appA-F | 247 | CAGATACGTCCAGTCCCGAT | |
| appA-R | AGTTCCGATGGTAATGCCTG | |||
| 16S rRNA | 338F | 468 | ACTCCTACGGGAGGCAGCAG | [ |
| 806R | GGACTACHVGGGTWTCTAAT |
表1 功能基因扩增引物
Table 1 Amplification primers for functional gene
| 引物Primer | 引物名称Primer name | 引物长度Primer length/bp | 引物序列Primer sequence(5'-3') | 参考文献Reference |
|---|---|---|---|---|
| phoD | ALPS-F730 | 370 | CAGTGGGACGA CCACGAGGT | [ |
| ALPS-1101 | GAGGCCGATCG GCATGTCG | |||
| phoC | phoC-A-F1 | 155 | CGGCTCCTATCCGTCCGG | [ |
| phoC-A-R1 | CAACATCGCTTTGCCAGTG | |||
| appA | appA-F | 247 | CAGATACGTCCAGTCCCGAT | |
| appA-R | AGTTCCGATGGTAATGCCTG | |||
| 16S rRNA | 338F | 468 | ACTCCTACGGGAGGCAGCAG | [ |
| 806R | GGACTACHVGGGTWTCTAAT |
| 引物Primer | 扩增体系Amplification system | 扩增程序Amplification procedure |
|---|---|---|
| phoD | 10 μL体系:Bestar Sybr Green qPCR Master Mix 5 μL,引物0.4 μL,DNA样品2 μL(10 ng·μL-1),灭菌水2.6 μL | 预变性95℃ 300 s,变性 95℃ 15 s,退火58℃ 30 s,延伸 72℃ 20 s,40 个循环 |
| phoC | 预变性 95℃ 300 s,变性 95℃ 15 s,退火57℃ 30 s,延伸 72℃ 20 s,40 个循环 | |
| appA | 预变性 95℃ 300 s,变性 95℃ 15 s,退火60℃ 30 s,延伸 72℃ 20 s,40 个循环 | |
| 16S rRNA | 预变性 95℃ 300 s,变性 95℃ 15 s,退火55℃ 30 s,延伸 72℃ 20 s,40 个循环 |
表2 荧光定量PCR反应条件
Table 2 Fluorescence quantitative PCR reaction conditions
| 引物Primer | 扩增体系Amplification system | 扩增程序Amplification procedure |
|---|---|---|
| phoD | 10 μL体系:Bestar Sybr Green qPCR Master Mix 5 μL,引物0.4 μL,DNA样品2 μL(10 ng·μL-1),灭菌水2.6 μL | 预变性95℃ 300 s,变性 95℃ 15 s,退火58℃ 30 s,延伸 72℃ 20 s,40 个循环 |
| phoC | 预变性 95℃ 300 s,变性 95℃ 15 s,退火57℃ 30 s,延伸 72℃ 20 s,40 个循环 | |
| appA | 预变性 95℃ 300 s,变性 95℃ 15 s,退火60℃ 30 s,延伸 72℃ 20 s,40 个循环 | |
| 16S rRNA | 预变性 95℃ 300 s,变性 95℃ 15 s,退火55℃ 30 s,延伸 72℃ 20 s,40 个循环 |
| 处理Treatment | 测定指标Measurement indicators | ||||||
|---|---|---|---|---|---|---|---|
| pH | 铵态氮 NH4+-N/(mg·kg-1) | 硝态氮 NO3--N/(mg·kg-1) | 有效磷 AP/(mg·kg-1) | 总磷 TP/(g·kg-1) | 总氮 TN/(g·kg-1) | 有机质 SOM/(g·kg-1) | |
| ZCK | 6.58±0.14 | 2.62±0.18 | 0.09±0.01 | 2.06±0.36 | 0.16±0.02 | 0.38±0.04 | 10.66±0.72 |
| ZN | 5.78±0.05** | 17.89±1.09** | 0.60±0.12* | 1.12±0.05* | 0.10±0.02* | 0.45±0.04 | 12.18±0.55 |
表3 氮添加对土壤基本理化性质的影响
Table 3 Effects of nitrogen addition on the basic physical and chemical properties of soil
| 处理Treatment | 测定指标Measurement indicators | ||||||
|---|---|---|---|---|---|---|---|
| pH | 铵态氮 NH4+-N/(mg·kg-1) | 硝态氮 NO3--N/(mg·kg-1) | 有效磷 AP/(mg·kg-1) | 总磷 TP/(g·kg-1) | 总氮 TN/(g·kg-1) | 有机质 SOM/(g·kg-1) | |
| ZCK | 6.58±0.14 | 2.62±0.18 | 0.09±0.01 | 2.06±0.36 | 0.16±0.02 | 0.38±0.04 | 10.66±0.72 |
| ZN | 5.78±0.05** | 17.89±1.09** | 0.60±0.12* | 1.12±0.05* | 0.10±0.02* | 0.45±0.04 | 12.18±0.55 |

图1 对照(ZCK)和氮添加(ZN)处理下土壤微生物量磷(MBP)和微生物量碳(MBC)的变化
Fig.1 Changes in soil microbial biomass phosphorus(MBP)and microbial biomass carbon(MBC)under control(ZCK)and nitrogen addition(ZN)treatments
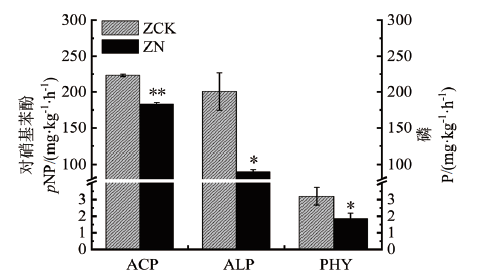
图2 对照(ZCK)和氮添加(ZN)处理下土壤有机磷转化的3种酶活性的变化
Fig. 2 Changes in activities of three enzymes involved in soil organic phosphorus conversion under control(ZCK)and nitrogen addition(ZN)treatments
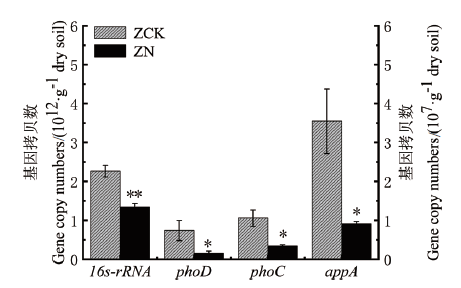
图3 对照(ZCK)和氮添加(ZN)处理下土壤16S rRNA、phoD、phoC、appA的基因拷贝数
Fig. 3 Gene copy numbers of soil 16S rRNA,phoD,phoC,and appA under control(ZCK)and nitrogen addition(ZN)treatments
| pH | MBP | MBC | NH4+-N | NO3--N | AP | TP | TN | SOM | 16S rRNA | phoD | phoC | appA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACP | 0.890** | 0.864** | 0.877** | -0.995** | -0.918** | 0.692 | 0.582 | -0.465 | -0.528 | 0.883** | 0.595 | 0.764* | 0.735* |
| ALP | 0.883** | 0.725* | 0.764* | -0.860** | -0.739* | 0.438 | 0.346 | -0.382 | -0.220 | 0.799* | 0.267 | 0.597 | 0.646 |
| PHY | 0.692 | 0.811* | 0.757* | -0.642 | -0.698 | 0.361 | 0.484 | -0.095 | -0.589 | 0.383 | 0.567 | 0.344 | 0.396 |
| 16S rRNA | 0.762* | 0.755* | 0.796* | -0.910** | -0.791* | 0.709* | 0.550 | -0.430 | -0.524 | 1 | 0.671 | 0.880** | 0.923** |
| phoD | 0.461 | 0.731* | 0.727* | -0.631 | -0.582 | 0.856** | 0.857 | -0.500 | -0.884** | 0.671 | 1 | 0.865** | 0.796* |
| phoC | 0.596 | 0.713* | 0.781* | -0.782* | -0.674 | 0.935** | 0.770 | -0.653 | -0.775* | 0.880** | 0.865** | 1 | 0.902** |
| appA | 0.676 | 0.772* | 0.821* | -0.781* | -0.698 | 0.702 | 0.608 | -0.464 | -0.677 | 0.923** | 0.796* | 0.902** | 1 |
表4 土壤酶活性、基因拷贝数、土壤理化性质间的相关性
Table 4 Correlation among soil enzyme activities,gene copy numbers and soil physicochemical properties
| pH | MBP | MBC | NH4+-N | NO3--N | AP | TP | TN | SOM | 16S rRNA | phoD | phoC | appA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACP | 0.890** | 0.864** | 0.877** | -0.995** | -0.918** | 0.692 | 0.582 | -0.465 | -0.528 | 0.883** | 0.595 | 0.764* | 0.735* |
| ALP | 0.883** | 0.725* | 0.764* | -0.860** | -0.739* | 0.438 | 0.346 | -0.382 | -0.220 | 0.799* | 0.267 | 0.597 | 0.646 |
| PHY | 0.692 | 0.811* | 0.757* | -0.642 | -0.698 | 0.361 | 0.484 | -0.095 | -0.589 | 0.383 | 0.567 | 0.344 | 0.396 |
| 16S rRNA | 0.762* | 0.755* | 0.796* | -0.910** | -0.791* | 0.709* | 0.550 | -0.430 | -0.524 | 1 | 0.671 | 0.880** | 0.923** |
| phoD | 0.461 | 0.731* | 0.727* | -0.631 | -0.582 | 0.856** | 0.857 | -0.500 | -0.884** | 0.671 | 1 | 0.865** | 0.796* |
| phoC | 0.596 | 0.713* | 0.781* | -0.782* | -0.674 | 0.935** | 0.770 | -0.653 | -0.775* | 0.880** | 0.865** | 1 | 0.902** |
| appA | 0.676 | 0.772* | 0.821* | -0.781* | -0.698 | 0.702 | 0.608 | -0.464 | -0.677 | 0.923** | 0.796* | 0.902** | 1 |
| [1] |
Attiwill PM, Adams MA. Nutrient cycling in forests[J]. New Phytol, 1993, 124(4):561-582.
doi: 10.1111/j.1469-8137.1993.tb03847.x pmid: 33874438 |
| [2] | 赵少华, 宇万太, 张璐, 等. 土壤有机磷研究进展[J]. 应用生态学报, 2004, 15(11):2189-2194. |
| Zhao SH, Yu WT, Zhang L, et al. Research advance in soil organic phosphorus[J]. Chin J Appl Ecol, 2004, 15(11):2189-2194. | |
| [3] | 李巧玲, 李爱博, 黄志远, 等. 解磷微生物在林业土壤生态修复中的应用进展[J]. 世界林业研究, 2022, 35(1):15-20. |
| Li QL, Li AB, Huang ZY, et al. Application of phosphorus solubilizing microorganisms in forestry soil ecological restoration[J]. World For Res, 2022, 35(1):15-20. | |
| [4] | 滕泽栋, 李敏, 朱静, 等. 解磷微生物对土壤磷资源利用影响的研究进展[J]. 土壤通报, 2017, 48(1):229-235. |
| Teng ZD, Li M, Zhu J, et al. Research advances in effect of phosphate-solubilizing microorganisms on soil phosphorus resource utilization[J]. Chin J Soil Sci, 2017, 48(1):229-235. | |
| [5] |
Dick WA, Cheng L, Wang P. Soil acid and alkaline phosphatase activity as pH adjustment indicators[J]. Soil Biol Biochem, 2000, 32(13):1915-1919.
doi: 10.1016/S0038-0717(00)00166-8 URL |
| [6] | 王小春, 梁新强. 生态环境中植酸酶种类及来源分析[J]. 环境生态学, 2020, 2(4):51-56. |
|
Wang XC, Liang XQ. Analysis of the types and sources of phytase in ecological environment[J]. Environ Ecol, 2020, 2(4):51-56.
doi: 10.1890/1540-9295(2004)002[0051:PL]2.0.CO;2 URL |
|
| [7] | 郑曼曼, 王超, 沈仁芳. 碳酸钙和根际作用对酸性红壤解磷微生物丰度的影响[J]. 土壤, 2020, 52(4):704-709. |
| Zheng MM, Wang C, Shen RF. Effects of calcium carbonate and rhizosphere on abundance of phosphate-solubilizing microorganisms in acidic red soil[J]. Soils, 2020, 52(4):704-709. | |
| [8] |
Canfield DE, Glazer AN, Falkowski PG. The evolution and future of Earth's nitrogen cycle[J]. Science, 2010, 330(6001):192-196.
doi: 10.1126/science.1186120 pmid: 20929768 |
| [9] |
Wu JP, Liu WF, Fan HB, et al. Asynchronous responses of soil microbial community and understory plant community to simulated nitrogen deposition in a subtropical forest[J]. Ecol Evol, 2013, 3(11):3895-3905.
doi: 10.1002/ece3.750 pmid: 24198947 |
| [10] |
Tibbett M, Sanders FE. Ectomycorrhizal symbiosis can enhance plant nutrition through improved access to discrete organic nutrient patches of high resource quality[J]. Ann Bot, 2002, 89(6):783-789.
doi: 10.1093/aob/mcf129 URL |
| [11] |
Ikoyi I, Egeter B, Chaves C, et al. Responses of soil microbiota and nematodes to application of organic and inorganic fertilizers in grassland columns[J]. Biol Fertil Soils, 2020, 56(5):647-662.
doi: 10.1007/s00374-020-01440-5 URL |
| [12] |
Luo GW, Ling N, Xue C, et al. Nitrogen-inputs regulate microbial functional and genetic resistance and resilience to drying-rewetting cycles, with implications for crop yields[J]. Plant Soil, 2019, 441(1/2):301-315.
doi: 10.1007/s11104-019-04120-y URL |
| [13] |
Chen XD, Jiang N, Condron LM, et al. Soil alkaline phosphatase activity and bacterial phoD gene abundance and diversity under long-term nitrogen and manure inputs[J]. Geoderma, 2019, 349:36-44.
doi: 10.1016/j.geoderma.2019.04.039 URL |
| [14] | 林贵刚, 赵琼, 赵蕾, 等. 林下植被去除与氮添加对樟子松人工林土壤化学和生物学性质的影响[J]. 应用生态学报, 2012, 23(5):1188-1194. |
| Lin GG, Zhao Q, Zhao L, et al. Effects of understory removal and nitrogen addition on the soil chemical and biological properties of Pinus sylvestris var. mongolica plantation in Keerqin Sandy Land[J]. Chin J Appl Ecol, 2012, 23(5):1188-1194. | |
| [15] | 朱芸芸, 李敏, 曲博, 等. 湿地植物根际土壤磷酸酶活性变化规律研究[J]. 环境科学与技术, 2016, 39(10):106-112. |
| Zhu YY, Li M, Qu B, et al. Research on the variations of phosphatase activity in rhizosphere soil of wetland plants[J]. Environ Sci Technol, 2016, 39(10):106-112. | |
| [16] |
Dassa J, Marck C, Boquet PL. The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2. 5 acid phosphatase and glucose-1-phosphatase[J]. J Bacteriol, 1990, 172(9):5497-5500.
pmid: 2168385 |
| [17] |
Fraser TD, Lynch DH, Gaiero J, et al. Quantification of bacterial non-specific acid(phoC)and alkaline(phoD)phosphatase genes in bulk and rhizosphere soil from organically managed soybean fields[J]. Appl Soil Ecol, 2017, 111:48-56.
doi: 10.1016/j.apsoil.2016.11.013 URL |
| [18] | 王玉荣, 杨成聪, 葛东颖, 等. 扩增区域对鲊广椒细菌MiSeq测序的影响[J]. 食品科学, 2019, 40(10):134-140. |
| Wang YR, Yang CC, Ge DY, et al. Influence of different amplified regions on results of bacterial diversity in zhaguangjiao, a Chinese traditional fermented chili product, by MiSeq sequencing[J]. Food Sci, 2019, 40(10):134-140. | |
| [19] | 杜荣骞. 生物统计学[M]. 北京: 高等教育出版社, 1999. |
| Du RQ. Biostatistics[M]. Beijing: Higher Education Press, 1999. | |
| [20] |
Waldrop MP, Zak DR, Sinsabaugh RL, et al. Nitrogen deposition modifies soil carbon storage through changes in microbial enzymatic activity[J]. Ecol Appl, 2004, 14(4):1172-1177.
doi: 10.1890/03-5120 URL |
| [21] |
Wei CZ, Yu Q, Bai E, et al. Nitrogen deposition weakens plant-microbe interactions in grassland ecosystems[J]. Glob Chang Biol, 2013, 19(12):3688-3697.
doi: 10.1111/gcb.12348 URL |
| [22] |
Wang C, Liu DW, Bai E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition[J]. Soil Biol Biochem, 2018, 120:126-133.
doi: 10.1016/j.soilbio.2018.02.003 URL |
| [23] |
Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria[J]. Ecology, 2007, 88(6):1354-1364.
pmid: 17601128 |
| [24] |
Tan H, Barret M, Mooij MJ, et al. Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phoD phosphorus mineraliser group in pasture soils[J]. Biol Fertil Soils, 2013, 49(6):661-672.
doi: 10.1007/s00374-012-0755-5 URL |
| [25] |
Chen XD, Jiang N, Chen ZH, et al. Response of soil phoD phosphatase gene to long-term combined applications of chemical fertilizers and organic materials[J]. Appl Soil Ecol, 2017, 119:197-204.
doi: 10.1016/j.apsoil.2017.06.019 URL |
| [26] |
Zhalnina K, Dias R, de Quadros PD, et al. Soil pH determines microbial diversity and composition in the park grass experiment[J]. Microb Ecol, 2015, 69(2):395-406.
doi: 10.1007/s00248-014-0530-2 pmid: 25395291 |
| [27] |
Taylor JP, Wilson B, Mills MS, et al. Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques[J]. Soil Biol Biochem, 2002, 34(3):387-401.
doi: 10.1016/S0038-0717(01)00199-7 URL |
| [28] |
Zheng MM, Wang C, Li WX, et al. Soil nutrients drive function and composition of phoC-harboring bacterial community in acidic soils of Southern China[J]. Front Microbiol, 2019, 10:2654.
doi: 10.3389/fmicb.2019.02654 pmid: 31824452 |
| [29] |
Ragot SA, Huguenin-Elie O, Kertesz MA, et al. Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil[J]. Plant Soil, 2016, 408(1/2):15-30.
doi: 10.1007/s11104-016-2902-5 URL |
| [30] |
Rana MS, Sun XC, Imran M, et al. Mo-inefficient wheat response toward molybdenum supply in terms of soil phosphorus availability[J]. J Soil Sci Plant Nutr, 2020, 20(3):1560-1573.
doi: 10.1007/s42729-020-00298-8 URL |
| [31] |
Jenkinson DS, Ladd JN. Microbial biomass in soil:measurement and turnover[J]. Soil biochemistry, 1981, 5(1):415-471.
doi: 10.1016/0038-0717(73)90068-0 URL |
| [32] |
Fraser TD, Lynch DH, Bent E, et al. Soil bacterial phoD gene abundance and expression in response to applied phosphorus and long-term management[J]. Soil Biol Biochem, 2015, 88:137-147.
doi: 10.1016/j.soilbio.2015.04.014 URL |
| [33] |
Recena R, Cade-Menun BJ, Delgado A. Organic phosphorus forms in agricultural soils under Mediterranean climate[J]. Soil Sci Soc Am J, 2018, 82(4):783-795.
doi: 10.2136/sssaj2017.10.0360 URL |
| [34] |
Nannipieri P, Giagnoni L, Renella G, et al. Soil enzymology:classical and molecular approaches[J]. Biol Fertil Soils, 2012, 48(7):743-762.
doi: 10.1007/s00374-012-0723-0 URL |
| [35] |
丁锐, 陈旭辉, 李炳学. 植酸酶研究进展及土壤植酸酶应用展望[J]. 生物技术通报, 2019, 35(7):190-195.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1027 |
|
Ding R, Chen XH, Li BX. Research advances on phytase and prospect of applying soil phytase[J]. Biotechnol Bull, 2019, 35(7):190-195.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1027 |
|
| [36] | Nannipieri P, Giagnoni L, Landi L, et al. Role of Phosphatase Enzymes in Soil[M]// Bünemann E., Oberson A., Frossard E. Phosphorus Action. Berlin:Springer, 2011:215-243. |
| [37] |
Neumann G, Römheld V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants[J]. Plant Soil, 1999, 211(1):121-130.
doi: 10.1023/A:1004380832118 URL |
| [38] |
Treseder KK, Vitousek PM. Effects of soil nutrient availability on investment in acquisition of N and P in Hawaiian rain forests[J]. Ecology, 2001, 82(4):946-954.
doi: 10.1890/0012-9658(2001)082[0946:EOSNAO]2.0.CO;2 URL |
| [1] | 李琦, 杨晓蕾, 李晓林, 申友磊, 李建宏, 姚拓. 高寒草地燕麦根际解植酸磷促生菌鉴定及其优势菌假单胞菌属菌株功能特性[J]. 生物技术通报, 2023, 39(3): 243-253. |
| [2] | 李雅楠, 余利红, 陈新美, 杨浩萌, 黄火清. 来源于Penicillium sp.C1的水产用中性植酸酶基因在毕赤酵母中的表达及性质研究[J]. 生物技术通报, 2020, 36(2): 134-141. |
| [3] | 丁锐, 陈旭辉, 李炳学. 植酸酶研究进展及土壤植酸酶应用展望[J]. 生物技术通报, 2019, 35(7): 190-195. |
| [4] | 袁林, 黄朝, 曾静, 郭建军, 张婷, 吕珺,. 植酸酶YiAPPA与生淀粉结合域SBD融合酶的构建及酶学性质分析[J]. 生物技术通报, 2018, 34(3): 200-207. |
| [5] | 黄大昉. 转基因农作物育种发展与思考[J]. 生物技术通报, 2015, 31(4): 3-6. |
| [6] | 李雅楠, 黄火清, 姚斌, 赵青, 刘昆, 马文康. 来源于假单孢菌206植酸酶基因的克隆、表达及酶学性质研究[J]. 生物技术通报, 2013, 0(8): 139-144. |
| [7] | 高启禹, 李宏彬, 陈红丽, 孔雨, 周晨妍. 壳聚糖修饰的PLGA纳米颗粒固定化碱性磷酸单酯酶的技术研究[J]. 生物技术通报, 2013, 0(5): 199-204. |
| [8] | 罗俊彩;扈进冬;吴远征;李晓龙;李纪顺;杨合同;. 侧耳木霉T2-1植酸酶基因的序列分析及蛋白结构预测[J]. , 2011, 0(04): 193-198. |
| [9] | 冯慧玲;李春梅;吴振芳;陈惠;. 易错PCR技术提高黑曲霉N25植酸酶活力的研究[J]. , 2010, 0(10): 226-230. |
| [10] | 张群;丁庆豹;欧伶;夏林根;. 麦芽根中5′-磷酸二酯酶和5′-磷酸单酯酶的提取[J]. , 2008, 0(S1): 378-383. |
| [11] | . 信息交流[J]. , 2007, 0(04): 9-63. |
| [12] | 张建社;刘明志;刘臻;. 植酸酶基因工程菌构建及其生产应用中问题分析[J]. , 2006, 0(05): 22-25. |
| [13] | 李健;彭远义;. 毕赤酵母Mut~+和Mut~s重组子表达植酸酶基因及其表达物的酶学性质比较[J]. , 2006, 0(03): 58-62. |
| [14] | . 国内信息[J]. , 2000, 0(04): 53-53. |
| [15] | 孙国凤. 用种子表达植酸酶,添加重组种子提高饲料效率[J]. , 1996, 0(04): 20-20. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||