








生物技术通报 ›› 2022, Vol. 38 ›› Issue (10): 124-131.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0069
唐跃辉( ), 赵雨凡, 林锦, 王胤, 曹博远, 车怡帆, 杨文杰, 包欣欣, 杨同文
), 赵雨凡, 林锦, 王胤, 曹博远, 车怡帆, 杨文杰, 包欣欣, 杨同文
收稿日期:2022-01-13
出版日期:2022-10-26
发布日期:2022-11-11
作者简介:唐跃辉,男,博士,副教授,研究方向:植物基因功能;E-mail:基金资助:
TANG Yue-hui( ), ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen
), ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen
Received:2022-01-13
Published:2022-10-26
Online:2022-11-11
摘要:
肽链释放因子RF1通过识别终止密码子在翻译终止过程中起重要调控作用。获得水稻苗期致死突变体,明确OseRF1-1表达模式,为进一步研究OseRF1-1功能奠定基础。以粳稻花之舞和T-DNA插入突变体为材料,利用Tail-PCR、石蜡切片技术、Southern杂交和Northern杂交技术鉴定一个肽链释放因子eRF1-1的T-DNA单拷贝插入突变体ls。采用DNAMAN6.0软件和SMART对OseRF1-1进行生物信息学分析,利用RT-qPCR方法分析OseRF1-1的组织特异性表达。通过PEG介导方法转化拟南芥,明确OseRF1-1的亚细胞定位。结果显示,ls突变体植株在3-5叶分蘖期迅速死亡;ls突变体叶鞘与分蘖节连接区域褐化,分蘖节越往下部位症状较严重;Tail-PCR分析结果显示,ls突变体T-DNA插入时丢失了T-DNA的右边界和NOS的终止子序列导致GUS和OseRF1-1融合转录,而GUS的终止密码子仍然存在,进而引起OseRF1-1不能进行正常翻译;ls突变体转录出GUS和OseRF1-1的融合转录子;OseRF1-1在各组织中均检测到表达且在穗中高表达;该基因定位于细胞核和细胞质。ls突变体导致水稻苗期致死可能是由于T-DNA插入OseRF1-1导致该基因不能正常转录引起。
唐跃辉, 赵雨凡, 林锦, 王胤, 曹博远, 车怡帆, 杨文杰, 包欣欣, 杨同文. 水稻苗期致死突变体的鉴定及其基因定位[J]. 生物技术通报, 2022, 38(10): 124-131.
TANG Yue-hui, ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen. Identification and Gene Mapping of a Seedling Lethal Mutant in Rice[J]. Biotechnology Bulletin, 2022, 38(10): 124-131.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Purpose |
|---|---|---|---|
| GUSF | GATGTTGGCGACCTCGTATT | 354 | 探针制备Probe preparation |
| GUSR | ACAGCGTCTCCGACCTGAT | ||
| eRF1-1F | GCTACTCCTACAAAACACCAGATG | 368 | |
| eRF1-1R | GCATACCAAGAAGGCAAGAAC | ||
| RAD1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA | Tail-PCR | |
| RAD2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT | ||
| RAD3 | ACGATGGACTCCAGAGCGGCCGCHNVNNNCCAC | ||
| RAD4 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA | ||
| RAD5 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT | ||
| AC | ACGATGGACTCCAGAG | ||
| RB1 | GTCGTCGGTGAACAGGTATGGAAT | ||
| RB2 | ACGATGGACTCCAGTCCGGCCTGGCGGTAACAAG- AAAGGGATCTTCACT | ||
| RB3 | AAACCGCAGCAGGGAGGCAAAC | ||
| OseRF1-1F | CGAGCACGACACGAGCTTT | 179 | 定量表达分析Quantitative expression analysis |
| OseRF1-1R | GTGCAGCATCAAGTCCTTTAAT | ||
| OseRF1-2F | AATCGTTCGTCCTGCTGCTT | 151 | |
| OseRF1-2R | TTGCCTCTAGCAGACTCCAGTG | ||
| OseRF1-3F | ACGAACATCACCGCAACCT | 178 | |
| OseRF1-3R | CGAAATCATGCTCGTACCGTT | ||
| OseRF1-4F | AAGTTCAGAGGCGAGGGTTT | 176 | |
| OseRF1-4R | CTGACTCCAGAGCCTTGATTAG | ||
| RUB1F | AGGGTTCACAAGTCTGCCTATT | 165 | |
| RUB1R | TTCCATGCTGCTCTACCACAG | ||
| OseRF1-1F OseRF1-1R | CGAGCACGACACGAGCTTT AGATCCAACGTCACAGGGTACAT | 1 342 | 亚细胞定位载体构建引物 Subcellular localization vector construction primers |
表1 本研究所需引物
Table 1 Primers used in the study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Purpose |
|---|---|---|---|
| GUSF | GATGTTGGCGACCTCGTATT | 354 | 探针制备Probe preparation |
| GUSR | ACAGCGTCTCCGACCTGAT | ||
| eRF1-1F | GCTACTCCTACAAAACACCAGATG | 368 | |
| eRF1-1R | GCATACCAAGAAGGCAAGAAC | ||
| RAD1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA | Tail-PCR | |
| RAD2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT | ||
| RAD3 | ACGATGGACTCCAGAGCGGCCGCHNVNNNCCAC | ||
| RAD4 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA | ||
| RAD5 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT | ||
| AC | ACGATGGACTCCAGAG | ||
| RB1 | GTCGTCGGTGAACAGGTATGGAAT | ||
| RB2 | ACGATGGACTCCAGTCCGGCCTGGCGGTAACAAG- AAAGGGATCTTCACT | ||
| RB3 | AAACCGCAGCAGGGAGGCAAAC | ||
| OseRF1-1F | CGAGCACGACACGAGCTTT | 179 | 定量表达分析Quantitative expression analysis |
| OseRF1-1R | GTGCAGCATCAAGTCCTTTAAT | ||
| OseRF1-2F | AATCGTTCGTCCTGCTGCTT | 151 | |
| OseRF1-2R | TTGCCTCTAGCAGACTCCAGTG | ||
| OseRF1-3F | ACGAACATCACCGCAACCT | 178 | |
| OseRF1-3R | CGAAATCATGCTCGTACCGTT | ||
| OseRF1-4F | AAGTTCAGAGGCGAGGGTTT | 176 | |
| OseRF1-4R | CTGACTCCAGAGCCTTGATTAG | ||
| RUB1F | AGGGTTCACAAGTCTGCCTATT | 165 | |
| RUB1R | TTCCATGCTGCTCTACCACAG | ||
| OseRF1-1F OseRF1-1R | CGAGCACGACACGAGCTTT AGATCCAACGTCACAGGGTACAT | 1 342 | 亚细胞定位载体构建引物 Subcellular localization vector construction primers |

图1 野生型和ls突变体表型 a:3-5叶期的ls突变体表型;b:野生型和ls突变体植株苗期茎基部和叶鞘表型
Fig. 1 Phenotype of wild type and ls mutant a:Phenotype of ls mutants at 3-5 leaf stage. b:Phenotype of stem base and sheath at seedling stage in wild type and ls mutant plants
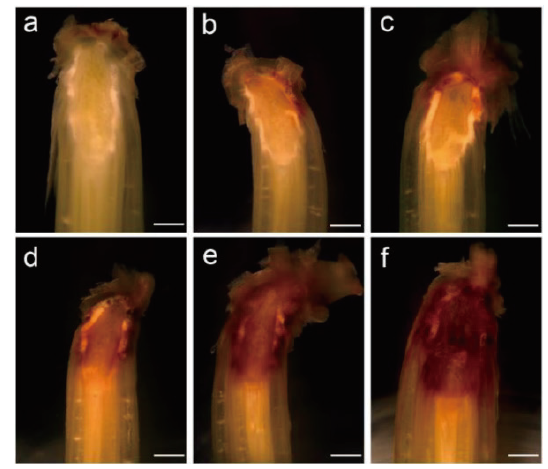
图2 野生型和ls突变体叶鞘与分蘖节部位显微镜结构分析 a:WT;b-f:ls突变体。Bar=1 mm
Fig. 2 Microscopic structure analysis of wild type and ls mutants leaf sheaths and tiller node sites a:WT;b-f:ls mutants. Bar=1 mm
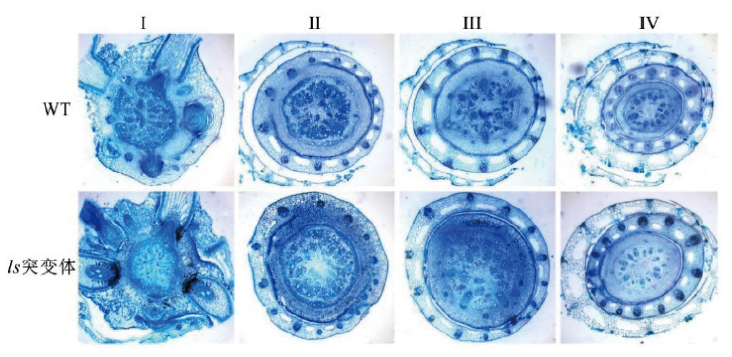
图3 野生型和ls突变体横切面结构 I-IV:野生型和ls突变体分蘖节从下往上横切面
Fig. 3 Cross-sections of wild-type and ls mutant I-IV:Cross-section of tiller nodes of wild type and ls mutants
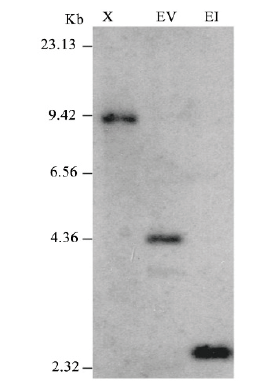
图4 ls突变体基因组DNA Southern杂交 以潮霉素基因序列为探针,X:Xba I;Ev:EcoR V;EI:EcoR I
Fig. 4 Southern blot of genome DNA in the ls mutant Hygromycin gene sequence was used as probe. X:Xba I;Ev:EcoR V;EI:EcoR I
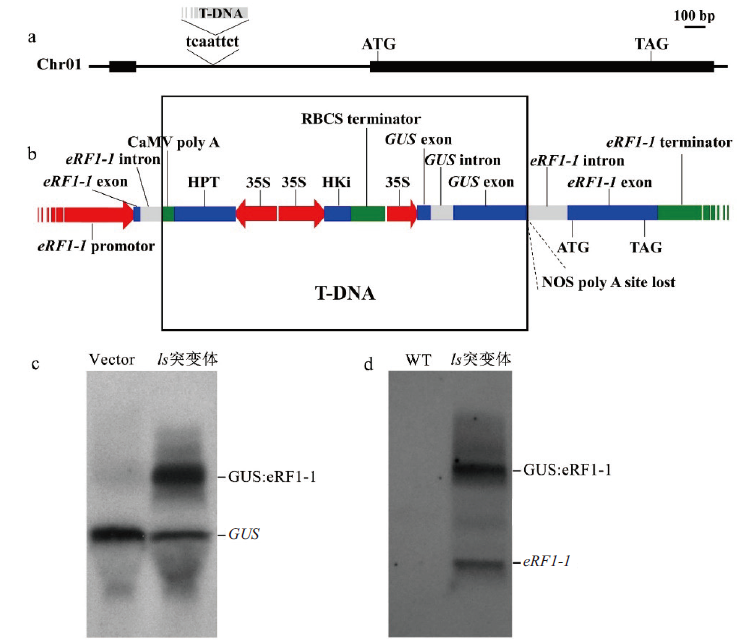
图5 ls突变体中T-DNA插入位置分析 a:T-DNA插入位点;b:T-DNA插入转录本分析;c:以GUS基因序列作为探针,Northern杂交;d:以eRF1-1基因序列作为探针,Northern杂交
Fig. 5 T-DNA insertion site analysis of ls mutant a:T-DNA insertion site;b:T-DNA insert transcript analysis;c:GUS gene sequence was used as probe. Northern hybridization;d:eRF1-1 gene sequence was used as probe. Northern hybridization

图6 eRF1和其同源蛋白氨基酸序列分析 OseRF1-1:水稻,Os01g0939500;OseRF1-2:水稻,XP_015630898;OseRF1-3:水稻,XP_025881251;OseRF1-4:水稻,XP_015647295;AteRF1-1:拟南芥,AT5G47880;AteRF1-2:拟南芥,AT1G12920;AteRF1-3:拟南芥,AT3G26618;TaeRF1:小麦,KAF6986654;ZmeRF1:玉米,ACG27998;AleRF1-3:玉山筷子芥,XP_002875327;BneRF1-3:中华菊头蝠,XP_019577167
Fig. 6 Analysis of amino acid sequences of eRF1 and its homologous proteins OseRF1-1:Oryza sativa Japonica,Os01g0939500;OseRF1-2:O. sativa,XP_015630898;OseRF1-3:O. sativa,XP_025881251;OseRF1-4:O. sativa,XP_015647295;AteRF1-1:Arabidopsis thaliana,AT5G47880;AteRF1-2:A. thaliana,AT1G12920;AteRF1-3:A. thaliana,AT3G26618;TaeRF1:Triticum aestivum,KAF6986654;ZmeRF1:Zea mays L.,ACG27998;AleRF1-3:Arabidopsis lyrata subsp. Lyrate,XP_002875327;BneRF1-3:Rhinolophus sinicus,XP_019577167
| [1] |
Mangkalaphiban K, He F, Ganesan R, et al. Transcriptome-wide investigation of stop codon readthrough in Saccharomyces cerevisiae[J]. PLoS Genet, 2021, 17(4):e1009538.
doi: 10.1371/journal.pgen.1009538 URL |
| [2] |
Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes—The impact of RNA sequence[J]. RNA Biol, 2015, 12(9):950-958.
doi: 10.1080/15476286.2015.1068497 pmid: 26176195 |
| [3] |
Crawford DJ, Ito K, Nakamura Y, et al. Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3[J]. EMBO J, 1999, 18(3):727-732.
pmid: 9927432 |
| [4] |
Saito K, Ito K. Genetic analysis of L123 of the tRNA-mimicking eukaryote release factor eRF1, an amino acid residue critical for discrimination of stop codons[J]. Nucleic Acids Res, 2015, 43(9):4591-4601.
doi: 10.1093/nar/gkv376 pmid: 25897120 |
| [5] |
Bertram G, Bell HA, Ritchie DW, et al. Terminating eukaryote translation:domain 1 of release factor eRF1 functions in stop codon recognition[J]. RNA, 2000, 6(9):1236-1247.
pmid: 10999601 |
| [6] |
Beiβel C, Neumann B, Uhse S, et al. Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3[J]. Nucleic Acids Res, 2019, 47(9):4798-4813.
doi: 10.1093/nar/gkz177 pmid: 30873535 |
| [7] |
Frolova LY, Tsivkovskii RY, Sivolobova GF, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis[J]. RNA, 1999, 5(8):1014-1020.
pmid: 10445876 |
| [8] |
Kurilla A, Szőke A, Auber A, et al. Expression of the translation termination factor eRF1 is autoregulated by translational readthrough and 3’UTR intron-mediated NMD in Neurospora crassa[J]. FEBS Lett, 2020, 594(21):3504-3517.
doi: 10.1002/1873-3468.13918 URL |
| [9] |
Nyikó T, Auber A, Szabadkai L, et al. Expression of the eRF1 translation termination factor is controlled by an autoregulatory circuit involving readthrough and nonsense-mediated decay in plants[J]. Nucleic Acids Res, 2017, 45(7):4174-4188.
doi: 10.1093/nar/gkw1303 pmid: 28062855 |
| [10] |
Yang Q, Yu CH, Zhao FZ, et al. eRF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons[J]. Nucleic Acids Res, 2019, 47(17):9243-9258.
doi: 10.1093/nar/gkz710 URL |
| [11] |
An Y, Lou YF, Xu YW. Overexpression, crystallization and preliminary X-ray crystallographic analysis of release factor eRF1-1 from Arabidopsis thaliana[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2013, 69(Pt 11):1295-1298.
doi: 10.1107/S1744309113027784 URL |
| [12] | Petsch KA, Mylne J, Botella JR. Cosuppression of eukaryotic release factor 1-1 in Arabidopsis affects cell elongation and radial cell division[J]. Plant Physiol, 2005, 139(1):115-126. |
| [13] |
Zhou XJ, Cooke P, Li L. Eukaryotic release factor 1-2 affects Arabidopsis responses to glucose and phytohormones during germination and early seedling development[J]. J Exp Bot, 2010, 61(2):357-367.
doi: 10.1093/jxb/erp308 URL |
| [14] |
Zhao B, Tang YY, Zhang BC, et al. The temperature-dependent retention of introns in GPI8 transcripts contributes to a drooping and fragile shoot phenotype in rice[J]. Int J Mol Sci, 2019, 21(1):299.
doi: 10.3390/ijms21010299 URL |
| [15] |
Codner GF, Erbs V, Loeffler J, et al. Universal Southern blot protocol with cold or radioactive probes for the validation of alleles obtained by homologous recombination[J]. Methods, 2021, 191:59-67.
doi: 10.1016/j.ymeth.2020.06.011 URL |
| [16] |
Yang C, Ma YM, He Y, et al. OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling[J]. Plant J, 2018, 93(3):489-501.
doi: 10.1111/tpj.13793 URL |
| [17] |
Zia MF, Flynt AS. Detection and verification of mammalian mirtrons by northern blotting[J]. Methods Mol Biol, 2018, 1823:209-219.
doi: 10.1007/978-1-4939-8624-8_16 pmid: 29959684 |
| [18] |
Tang YH, Li H, Guan YX, et al. Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5[J]. Front Genet, 2020, 11:670.
doi: 10.3389/fgene.2020.00670 URL |
| [19] |
Hellen CUT. Translation termination and ribosome recycling in eukaryotes[J]. Cold Spring Harb Perspect Biol, 2018, 10(10):a032656.
doi: 10.1101/cshperspect.a032656 URL |
| [20] |
Song H, Mugnier P, Das AK, et al. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis[J]. Cell, 2000, 100(3):311-321.
pmid: 10676813 |
| [21] |
Baradaran-Heravi A, Balgi AD, Hosseini-Farahabadi S, et al. Effect of small molecule eRF3 degraders on premature termination codon readthrough[J]. Nucleic Acids Res, 2021, 49(7):3692-3708.
doi: 10.1093/nar/gkab194 pmid: 33764477 |
| [22] |
Castellanos M, Mothi N, Muñoz V. Eukaryotic transcription factors can track and control their target genes using DNA antennas[J]. Nat Commun, 2020, 11(1):540.
doi: 10.1038/s41467-019-14217-8 pmid: 31992709 |
| [23] | 熊伟, 朱成新, 王玉婷, 等. CRISPR-Cas9介导靶向突变拟南芥ERF1-1基因[J]. 深圳大学学报:理工版, 2021, 38(5):504-509. |
| Xiong W, Zhu CX, Wang YT, et al. Targeted mutation of ERF1-1 gene in Arabidopsis using CRISPR-Cas9 gene editing system[J]. J Shenzhen Univ Sci Eng, 2021, 38(5):504-509. | |
| [24] |
Chen WC, Wang Q, Cao TJ, et al. UBC19 is a new interacting protein of ORANGE for its nuclear localization in Arabidopsis thaliana[J]. Plant Signal Behav, 2021, 16(11):1964847.
doi: 10.1080/15592324.2021.1964847 URL |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [3] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [4] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [5] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [6] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [7] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [8] | 李怡君, 吴晨晨, 李睿, 王喆, 何山文, 韦善君, 张晓霞. 水稻内生细菌新资源分离培养方案探究[J]. 生物技术通报, 2023, 39(4): 201-211. |
| [9] | 卢振万, 李雪琪, 黄金光, 周焕斌. 利用胞嘧啶碱基编辑技术创制耐草甘膦水稻[J]. 生物技术通报, 2023, 39(2): 63-69. |
| [10] | 杨茂, 林宇丰, 戴阳朔, 潘素君, 彭伟业, 严明雄, 李魏, 王冰, 戴良英. OsDIS1通过抗氧化途径负调控水稻耐旱性[J]. 生物技术通报, 2023, 39(2): 88-95. |
| [11] | 蒋铭轩, 李康, 罗亮, 刘建祥, 芦海平. 植物表达外源蛋白研究进展及展望[J]. 生物技术通报, 2023, 39(11): 110-122. |
| [12] | 姜南, 石杨, 赵志慧, 李斌, 赵熠辉, 杨俊彪, 闫家铭, 靳雨璠, 陈稷, 黄进. 镉胁迫下水稻OsPT1的表达及功能分析[J]. 生物技术通报, 2023, 39(1): 166-174. |
| [13] | 陈光, 李佳, 杜瑞英, 王旭. 水稻盐敏感突变体ss2的鉴定与基因功能分析[J]. 生物技术通报, 2022, 38(9): 158-166. |
| [14] | 高晓蓉, 丁尧, 吕军. 芘降解菌Pseudomonas sp. PR3的植物促生特性及其对芘胁迫下水稻生长的影响[J]. 生物技术通报, 2022, 38(9): 226-236. |
| [15] | 黄婧, 朱亮, 薛蓬勃, 付强. 水稻叶和籽粒镉积累机制及QTL定位研究[J]. 生物技术通报, 2022, 38(8): 118-126. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||