








生物技术通报 ›› 2023, Vol. 39 ›› Issue (8): 173-184.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0386
吴元明1( ), 林佳怡1, 柳雨汐1, 李丹婷2, 张宗琼2, 郑晓明3,4,5, 逄洪波1(
), 林佳怡1, 柳雨汐1, 李丹婷2, 张宗琼2, 郑晓明3,4,5, 逄洪波1( )
)
收稿日期:2023-04-22
出版日期:2023-08-26
发布日期:2023-09-05
通讯作者:
逄洪波,女,博士,副教授,研究方向:植物逆境分子生物学;E-mail: panghb@synu.edu.cn作者简介:吴元明,男,硕士研究生,研究方向:生物化学与分子生物学;E-mail: 19861602091@163.com
基金资助:
WU Yuan-ming1( ), LIN Jia-yi1, LIU Yu-xi1, LI Dan-ting2, ZHANG Zong-qiong2, ZHENG Xiao-ming3,4,5, PANG Hong-bo1(
), LIN Jia-yi1, LIU Yu-xi1, LI Dan-ting2, ZHANG Zong-qiong2, ZHENG Xiao-ming3,4,5, PANG Hong-bo1( )
)
Received:2023-04-22
Published:2023-08-26
Online:2023-09-05
摘要:
株高是影响水稻产量稳定性的重要因素之一,水稻株高相关QTL的定位及候选基因的挖掘,有助于深入了解株高分子调控机制,进而为培育理想株型的水稻品种奠定基础。以油占8号及广西野生稻为亲本构建的285个CSSL群体为试验对象,结合NGS与BSA-seq等方法,利用SNP和InDel 两种分子标记进行关联分析,对可能与株高相关的基因组区段进行定位。结果显示,Δ(SNP-index)分子标记在Chr.7和Chr.10中分别关联到3.205和1.311 Mb大小的候选基因组区域。Δ(InDel-index)标记关联到的基因组区域大小分别为2.848和1.292 Mb,且全部包含于Δ(SNP-index)关联到的区间内。结合GO、KEGG、Uniprot、eggNOG等数据库中的功能注释、高质量多态性位点筛选以及已有的株高相关转录组数据,最终关联到Chr.7中的5个候选基因,包括功能和机理已知的OsTCP21。RT-qPCR结果显示,OsTCP21在高株和矮株中的表达差异与已有研究结果相一致,LOC_Os07g05050和LOC_Os07g02850在高株中表达量较高,LOC_Os07g04220和LOC_Os07g02770在矮株中表达量较高。这5个候选基因在水稻株高性状的调控中起重要的作用,OsTCP21是一个关键调控基因。
吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184.
WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq[J]. Biotechnology Bulletin, 2023, 39(8): 173-184.
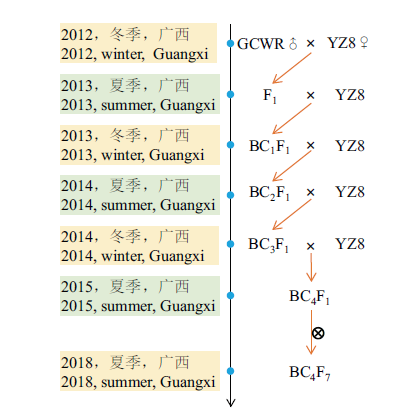
图1 本研究所用的回交群体的构建过程 GCWR:广西野生稻;YZ8:油占8号
Fig. 1 Construction process of the backcross populations used in this study GCWR: Guangxi Common Wild Rice; YZ8: Youzhan8
| 基因 Gene | 引物序列 Primer sequence(5'-3') | 产物大小 Product size/bp |
|---|---|---|
| LOC_Os07g02770 | F: GAGAGCATGGCAAGGAAGGT | 130 |
| R: TAGTGCGGTGCATTTGTCCT | ||
| LOC_Os07g02850 | F: GCGTCTGCAGGAAGAAGTCC | 125 |
| R: CTGCACCAGCCACATGAATC | ||
| LOC_Os07g04220 | F: CCCGCCATTAGTCCACAGAG | 128 |
| R: TTGCTGGTGGGCCTGATATG | ||
| LOC_Os07g05050 | F: CGAACTCCCAAACACCTCGG | 241 |
| R: GGTTCTTGGGCTTCCAGCTAA | ||
| LOC_Os07g05720 | F: TCTCCATGTCCTCGGGTCTT | 175 |
| R: GGGCTGAAGAGGAACGGAAT | ||
| OsActin | F: AGACCTTCAACACCCCTGCT | 110 |
| R: GTGGCTGACACCATCACCAG |
表1 用于RT-qPCR分析的引物列表
Table 1 List of primers for RT-qPCR analysis
| 基因 Gene | 引物序列 Primer sequence(5'-3') | 产物大小 Product size/bp |
|---|---|---|
| LOC_Os07g02770 | F: GAGAGCATGGCAAGGAAGGT | 130 |
| R: TAGTGCGGTGCATTTGTCCT | ||
| LOC_Os07g02850 | F: GCGTCTGCAGGAAGAAGTCC | 125 |
| R: CTGCACCAGCCACATGAATC | ||
| LOC_Os07g04220 | F: CCCGCCATTAGTCCACAGAG | 128 |
| R: TTGCTGGTGGGCCTGATATG | ||
| LOC_Os07g05050 | F: CGAACTCCCAAACACCTCGG | 241 |
| R: GGTTCTTGGGCTTCCAGCTAA | ||
| LOC_Os07g05720 | F: TCTCCATGTCCTCGGGTCTT | 175 |
| R: GGGCTGAAGAGGAACGGAAT | ||
| OsActin | F: AGACCTTCAACACCCCTGCT | 110 |
| R: GTGGCTGACACCATCACCAG |

图2 CSSL群体的株高表型分析 A:CSSL群体中最高株和最矮株表型(Bar=25 cm);B:CSSL群体平均株高的分布频率
Fig. 2 Plant height phenotype analysis of CSSL populations A: The tallest and shortest individuals in CSSL populations(Bar=25 cm).B: Distribution of average plant heights of CSSL populations
| 样品Sample | 原始数据 Raw base/bp | 过滤后的有效数据 Clean base/bp | 比对率Mapping rate/% | 平均测序深度Average depth(×) | 至少覆盖1个碱基的位点的比例Coverage at least 1×/% | 至少覆盖4个碱基的位点的比例Coverage at least 4×/% | Phred 数值大于20的碱基比例Q20/% | Phred 数值大于30的碱基比例Q30/% | 碱基G和C的含量GC/% |
|---|---|---|---|---|---|---|---|---|---|
| YZ8 | 11 134 196 100 | 11 134 054 800 | 99.00 | 23.81 | 82.46 | 79.06 | 97.57 | 93.31 | 44.60 |
| GCWR | 14 338 273 200 | 14 338 088 700 | 98.65 | 28.82 | 85.26 | 82.67 | 97.78 | 93.79 | 44.23 |
| H-pool | 5 067 688 200 | 5 067 637 200 | 99.04 | 11.36 | 82.27 | 74.08 | 97.11 | 92.15 | 43.59 |
| L-pool | 4 969 848 300 | 4 969 806 000 | 99.02 | 11.43 | 81.81 | 74.14 | 97.14 | 92.13 | 43.61 |
表2 用于BSA-seq分析的NGS测序数据统计概况
Table 2 Summary of NGS data for BSA-seq analysis
| 样品Sample | 原始数据 Raw base/bp | 过滤后的有效数据 Clean base/bp | 比对率Mapping rate/% | 平均测序深度Average depth(×) | 至少覆盖1个碱基的位点的比例Coverage at least 1×/% | 至少覆盖4个碱基的位点的比例Coverage at least 4×/% | Phred 数值大于20的碱基比例Q20/% | Phred 数值大于30的碱基比例Q30/% | 碱基G和C的含量GC/% |
|---|---|---|---|---|---|---|---|---|---|
| YZ8 | 11 134 196 100 | 11 134 054 800 | 99.00 | 23.81 | 82.46 | 79.06 | 97.57 | 93.31 | 44.60 |
| GCWR | 14 338 273 200 | 14 338 088 700 | 98.65 | 28.82 | 85.26 | 82.67 | 97.78 | 93.79 | 44.23 |
| H-pool | 5 067 688 200 | 5 067 637 200 | 99.04 | 11.36 | 82.27 | 74.08 | 97.11 | 92.15 | 43.59 |
| L-pool | 4 969 848 300 | 4 969 806 000 | 99.02 | 11.43 | 81.81 | 74.14 | 97.14 | 92.13 | 43.61 |

图3 基于BSA-seq分析的矮株(A)和高株(B)的SNP-index图以及Δ(SNP-index)图(C)
Fig. 3 SNP-index graphs of bulk-L(A)and bulk-H(B), as well as Δ(SNP-index)graph(C)based on BSA-seq analysis
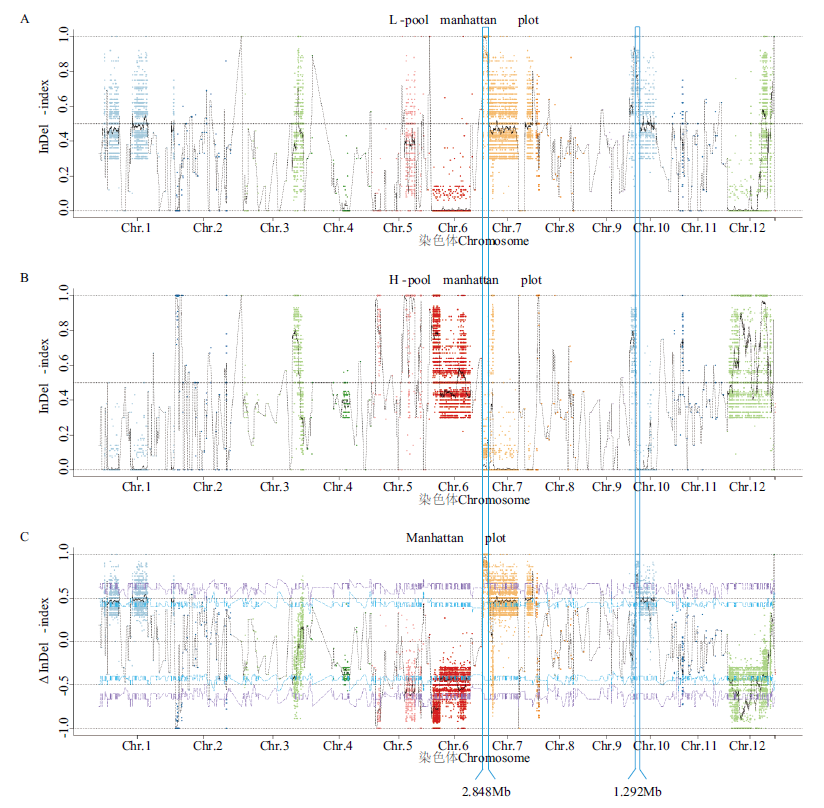
图4 基于BSA-seq分析的矮株(A)和高株(B)的InDel-index图以及Δ(InDel-index)图(C)
Fig. 4 InDel-index graphs of bulk-L(A)and bulk-H(B), as well as Δ(InDel -index)graph(C)based on BSA-seq analysis
| 染色体 Chromosome | 起始位置 Start/bp | 终止位置 End/bp | 大小 Size/Mb | 基因数量 Gene number |
|---|---|---|---|---|
| 7 | 1 | 2 848 000 | 2.848 | 141 |
| 10 | 3 796 001 | 5 088 000 | 1.292 | 135 |
| 总计Total | - | - | 4.140 | 276 |
表3 Δ(SNP-index)和Δ(InDel-index)定位与株高相关的候选基因组区段
Table 3 Candidate genome regions related to plant height mapping by Δ(SNP-index)and Δ(InDel-index)
| 染色体 Chromosome | 起始位置 Start/bp | 终止位置 End/bp | 大小 Size/Mb | 基因数量 Gene number |
|---|---|---|---|---|
| 7 | 1 | 2 848 000 | 2.848 | 141 |
| 10 | 3 796 001 | 5 088 000 | 1.292 | 135 |
| 总计Total | - | - | 4.140 | 276 |
| 功能注释数据库 Annotated database | 注释基因数目 Number of annotated genes |
|---|---|
| eggNOG | 26 |
| Uniprot | 41 |
| GO | 48 |
| KEGG | 20 |
| 总计Total | 78 |
表4 候选区间内基因的功能注释结果统计
Table 4 Statistics of functional annotation results for genes in candidate intervals
| 功能注释数据库 Annotated database | 注释基因数目 Number of annotated genes |
|---|---|
| eggNOG | 26 |
| Uniprot | 41 |
| GO | 48 |
| KEGG | 20 |
| 总计Total | 78 |
| 基因 Gene ID | 基因注释 Gene annotation |
|---|---|
| LOC_Os07g02770 | 含F-box和亮氨酸富集重复序列的蛋白 Protein containing F-box and leucine-enriched repeats |
| LOC_Os07g02850 | 含DUF538结构域的蛋白 Protein containing DUF538 domain |
| LOC_Os07g04220 | 光敏色素信号与损伤相关的受体类激酶 Wound and phytochrome signaling involved receptor like kinase |
| LOC_Os07g05050 | 含DEAD-box的ATP依赖的RNA解旋酶 ATP-dependent RNA helicase containing DEAD-box |
| LOC_Os07g05720 | TCP转录因子 TCP family transcription factor |
表5 候选基因功能注释结果
Table 5 Results of functional annotation of candidate genes
| 基因 Gene ID | 基因注释 Gene annotation |
|---|---|
| LOC_Os07g02770 | 含F-box和亮氨酸富集重复序列的蛋白 Protein containing F-box and leucine-enriched repeats |
| LOC_Os07g02850 | 含DUF538结构域的蛋白 Protein containing DUF538 domain |
| LOC_Os07g04220 | 光敏色素信号与损伤相关的受体类激酶 Wound and phytochrome signaling involved receptor like kinase |
| LOC_Os07g05050 | 含DEAD-box的ATP依赖的RNA解旋酶 ATP-dependent RNA helicase containing DEAD-box |
| LOC_Os07g05720 | TCP转录因子 TCP family transcription factor |
| [1] |
Spielmeyer W, Ellis MH, Chandler PM. Semidwarf(sd-1), green revolution rice, contains a defective gibberellin 20-oxidase gene[J]. Proc Natl Acad Sci USA, 2002, 99(13): 9043-9048.
doi: 10.1073/pnas.132266399 pmid: 12077303 |
| [2] |
Hedden P. The genes of the green revolution[J]. Trends Genet, 2003, 19(1): 5-9.
doi: 10.1016/s0168-9525(02)00009-4 pmid: 12493241 |
| [3] |
Sasaki A, Ashikari M, Ueguchi-Tanaka M, et al. Green revolution: a mutant gibberellin-synthesis gene in rice[J]. Nature, 2002, 416(6882): 701-702.
doi: 10.1038/416701a |
| [4] |
Peng J, Richards DE, Hartley NM, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators[J]. Nature, 1999, 400(6741): 256-261.
doi: 10.1038/22307 |
| [5] |
Liu C, Zheng S, Gui JS, et al. Shortened basal internodes encodes a gibberellin 2-oxidase and contributes to lodging resistance in rice[J]. Mol Plant, 2018, 11(2): 288-299.
doi: 10.1016/j.molp.2017.12.004 URL |
| [6] |
Tu B, Tao Z, Wang SG, et al. Loss of Gn1a/OsCKX2 confers heavy-panicle rice with excellent lodging resistance[J]. J Integr Plant Biol, 2022, 64(1): 23-38.
doi: 10.1111/jipb.v64.1 URL |
| [7] |
Ookawa T, Hobo T, Yano M, et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield[J]. Nat Commun, 2010, 1: 132.
pmid: 21119645 |
| [8] | Zegeye WA, Chen DB, Islam M, et al. OsFBK4, a novel GA insensitive gene positively regulates plant height in rice(Oryza Sativa L.)[J]. Ecol Genet Genom, 2022, 23: 100115. |
| [9] |
Liu TZ, Zhang X, Zhang H, et al. Dwarf and High Tillering1 represses rice tillering through mediating the splicing of D14 pre-mRNA[J]. Plant Cell, 2022, 34(9): 3301-3318.
doi: 10.1093/plcell/koac169 URL |
| [10] |
Hill WG. Understanding and using quantitative genetic variation[J]. Phil Trans R Soc B, 2010, 365(1537): 73-85.
doi: 10.1098/rstb.2009.0203 URL |
| [11] |
El-Soda M, Malosetti M, Zwaan BJ, et al. Genotype × environment interaction QTL mapping in plants: lessons from Arabidopsis[J]. Trends Plant Sci, 2014, 19(6): 390-398.
doi: 10.1016/j.tplants.2014.01.001 URL |
| [12] |
Hong Z, Ueguchi-Tanaka M, Umemura K, et al. A rice brassinosteroid-deficient mutant, ebisu dwarf(d2), is caused by a loss of function of a new member of cytochrome P450[J]. Plant Cell, 2003, 15(12): 2900-2910.
doi: 10.1105/tpc.014712 pmid: 14615594 |
| [13] |
Yamamuro C, Ihara Y, Wu X, et al. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint[J]. Plant Cell, 2000, 12(9): 1591-1606.
doi: 10.1105/tpc.12.9.1591 pmid: 11006334 |
| [14] |
Lin H, Wang RX, Qian Q, et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth[J]. Plant Cell, 2009, 21(5): 1512-1525.
doi: 10.1105/tpc.109.065987 pmid: 19470589 |
| [15] |
Multani DS, Briggs SP, Chamberlin MA, et al. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants[J]. Science, 2003, 302(5642): 81-84.
doi: 10.1126/science.1086072 pmid: 14526073 |
| [16] |
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760.
doi: 10.1093/bioinformatics/btp324 pmid: 19451168 |
| [17] |
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079.
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [18] |
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Res, 2010, 20(9): 1297-1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [19] |
Wang K, Li MY, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data[J]. Nucleic Acids Res, 2010, 38(16): e164.
doi: 10.1093/nar/gkq603 URL |
| [20] |
Takagi H, Abe A, Yoshida K, et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations[J]. Plant J, 2013, 74(1): 174-183.
doi: 10.1111/tpj.2013.74.issue-1 URL |
| [21] |
Sakai H, Lee SS, Tanaka T, et al. Rice annotation project database(RAP-DB): An integrative and interactive database for rice genomics[J]. Plant Cell Physiol, 2013, 54(2): e6.
doi: 10.1093/pcp/pcs183 URL |
| [22] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2-ΔΔCtmethod[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [24] |
Zou C, Wang PX, Xu YB. Bulked sample analysis in genetics, genomics and crop improvement[J]. Plant Biotechnol J, 2016, 14(10): 1941-1955.
doi: 10.1111/pbi.12559 pmid: 26990124 |
| [25] |
Xue HB, Shi T, Wang FF, et al. Interval mapping for red/green skin color in Asian pears using a modified QTL-seq method[J]. Hortic Res, 2017, 4: 17053.
doi: 10.1038/hortres.2017.53 URL |
| [26] |
Fu W, Huang SN, Gao Y, et al. Role of BrSDG8 on bolting in Chinese cabbage(Brassica rapa)[J]. Theor Appl Genet, 2020, 133(10): 2937-2948.
doi: 10.1007/s00122-020-03647-4 |
| [27] |
Zhang KJ, Li Y, Zhu WW, et al. Fine mapping and transcriptome analysis of virescent leaf gene v-2 in cucumber(Cucumis sativus L.)[J]. Front Plant Sci, 2020, 11: 570817.
doi: 10.3389/fpls.2020.570817 URL |
| [28] |
Zhang YX, Qin G, Ma QQ, et al. Identification of major locus Bph35 resistance to brown planthopper in rice[J]. Rice Sci, 2020, 27(3): 237-245.
doi: 10.1016/j.rsci.2020.04.006 URL |
| [29] |
Li ZQ, Xu YH. Bulk segregation analysis in the NGS era: a review of its teenage years[J]. Plant J, 2022, 109(6): 1355-1374.
doi: 10.1111/tpj.v109.6 URL |
| [30] |
Yang LM, Lei L, Li P, et al. Identification of candidate genes conferring cold tolerance to rice(Oryza sativa L.) at the bud-bursting stage using bulk segregant analysis sequencing and linkage mapping[J]. Front Plant Sci, 2021, 12: 647239.
doi: 10.3389/fpls.2021.647239 URL |
| [31] |
Liu G, Zhao TT, You XQ, et al. Molecular mapping of the Cf-10 gene by combining SNP/InDel-index and linkage analysis in tomato(Solanum lycopersicum)[J]. BMC Plant Biol, 2019, 19(1): 15.
doi: 10.1186/s12870-018-1616-7 |
| [32] |
Singh VK, Khan AW, Saxena RK, et al. Indel-seq: a fast-forward genetics approach for identification of trait-associated putative candidate genomic regions and its application in pigeonpea(Cajanus cajan)[J]. Plant Biotechnol J, 2017, 15(7): 906-914.
doi: 10.1111/pbi.12685 URL |
| [33] |
Kage U, Kumar A, Dhokane D, et al. Functional molecular markers for crop improvement[J]. Crit Rev Biotechnol, 2016, 36(5): 917-930.
doi: 10.3109/07388551.2015.1062743 pmid: 26171816 |
| [34] |
Wang RN, Yang XY, Guo S, et al. MiR319-targeted OsTCP21 and OsGAmyb regulate tillering and grain yield in rice[J]. J Integr Plant Biol, 2021, 63(7): 1260-1272.
doi: 10.1111/jipb.v63.7 URL |
| [35] |
Wang YH, Li JY. Rice, rising[J]. Nat Genet, 2008, 40(11): 1273-1275.
doi: 10.1038/ng1108-1273 pmid: 18957983 |
| [36] |
Tan LB, Li XR, Liu FX, et al. Control of a key transition from prostrate to erect growth in rice domestication[J]. Nat Genet, 2008, 40(11): 1360-1364.
doi: 10.1038/ng.197 pmid: 18820699 |
| [37] |
Jin J, Huang W, Gao JP, et al. Genetic control of rice plant architecture under domestication[J]. Nat Genet, 2008, 40(11): 1365-1369.
doi: 10.1038/ng.247 pmid: 18820696 |
| [38] |
Lan DY, Cao LM, Liu MY, et al. The identification and characterization of a plant height and grain length related gene hfr131 in rice[J]. Front Plant Sci, 2023, 14: 1152196.
doi: 10.3389/fpls.2023.1152196 URL |
| [39] |
Li Y, He YZ, Liu ZX, et al. OsSPL14 acts upstream of OsPIN1b and PILS6b to modulate axillary bud outgrowth by fine-tuning auxin transport in rice[J]. Plant J, 2022, 111(4): 1167-1182.
doi: 10.1111/tpj.v111.4 URL |
| [40] |
Zhang WF, Tan LB, Sun HY, et al. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice[J]. Mol Plant, 2019, 12(8): 1075-1089.
doi: 10.1016/j.molp.2019.04.005 URL |
| [41] |
Ding CH, Lin XH, Zuo Y, et al. Transcription factor OsbZIP49 controls tiller angle and plant architecture through the induction of indole-3-acetic acid-amido synthetases in rice[J]. Plant J, 2021, 108(5): 1346-1364.
doi: 10.1111/tpj.v108.5 URL |
| [42] |
Li Y, Li TT, He XR, et al. Blocking Osa-miR1871 enhances rice resistance against Magnaporthe oryzae and yield[J]. Plant Biotechnol J, 2022, 20(4): 646-659.
doi: 10.1111/pbi.v20.4 URL |
| [43] |
Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships[J]. Curr Opin Struct Biol, 1993, 3(3): 419-429.
doi: 10.1016/S0959-440X(05)80116-2 URL |
| [44] |
Gong ZZ, Lee H, Xiong LM, et al. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance[J]. Proc Natl Acad Sci USA, 2002, 99(17): 11507-11512.
doi: 10.1073/pnas.172399299 pmid: 12165572 |
| [45] |
Wang D, Qin BX, Li X, et al. Nucleolar DEAD-box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice[J]. PLoS Genet, 2016, 12(2): e1005844.
doi: 10.1371/journal.pgen.1005844 URL |
| [46] |
Yang SD, Guo DL, Pei MS, et al. Identification of the DEAD-box RNA helicase family members in grapevine reveals that VviDEADRH25a confers tolerance to drought stress[J]. J Integr Agric, 2022, 21(5): 1357-1374.
doi: 10.1016/S2095-3119(21)63870-4 URL |
| [47] | Takahashi S, Yoshikawa M, Kamada A, et al. The photoconvertible water-soluble chlorophyll-binding protein of Chenopodium album is a member of DUF538, a superfamily that distributes in Embryophyta[J]. J Plant Physiol, 2013, 170(17): 1549-1552. |
| [48] |
Gholizadeh A. DUF538 protein superfamily is predicted to be chlorophyll hydrolyzing enzymes in plants[J]. Physiol Mol Biol Plants, 2016, 22(1): 77-85.
doi: 10.1007/s12298-015-0331-1 URL |
| [49] |
Li QQ, Wu GX, Zhao YP, et al. CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height[J]. Plant Biotechnol J, 2020, 18(12): 2520-2532.
doi: 10.1111/pbi.v18.12 URL |
| [50] |
Li B, Du X, Fei YY, et al. Efficient breeding of early-maturing rice cultivar by editing PHYC via CRISPR/Cas9[J]. Rice, 2021, 14(1): 86.
doi: 10.1186/s12284-021-00527-3 |
| [51] | Kipreos ET, Pagano M. The F-box protein family[J]. Genome Biol, 2000, 1(5): reviews3002.1. |
| [52] |
Yu K, Yang WQ, Zhao B, et al. The Kelch-F-box protein Small and Glossy Leaves 1(SAGL1)negatively influences salicylic acid biosynthesis in Arabidopsis thaliana by promoting the turn-over of transcription factor Systemic Acquired Resistance Deficient 1(SARD1)[J]. New Phytol, 2022, 235(3): 885-897.
doi: 10.1111/nph.18197 pmid: 35491444 |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [3] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [4] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [5] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [6] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [7] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [8] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [9] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [10] | 徐建霞, 丁延庆, 冯周, 曹宁, 程斌, 高旭, 邹桂花, 张立异. 基于Super-GBS的高粱株高和节间数QTL定位[J]. 生物技术通报, 2023, 39(7): 185-194. |
| [11] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [12] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [13] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [14] | 李怡君, 吴晨晨, 李睿, 王喆, 何山文, 韦善君, 张晓霞. 水稻内生细菌新资源分离培养方案探究[J]. 生物技术通报, 2023, 39(4): 201-211. |
| [15] | 卢振万, 李雪琪, 黄金光, 周焕斌. 利用胞嘧啶碱基编辑技术创制耐草甘膦水稻[J]. 生物技术通报, 2023, 39(2): 63-69. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||