








生物技术通报 ›› 2023, Vol. 39 ›› Issue (5): 177-191.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0951
刘辉( ), 卢扬(
), 卢扬( ), 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发
), 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发
收稿日期:2022-07-31
出版日期:2023-05-26
发布日期:2023-06-08
通讯作者:
卢扬,男,硕士,副研究员,研究方向:作物栽培与育种;E-mail: 499528997@qq.com作者简介:刘辉,男,博士,副研究员,研究方向:生物化学与分子生物学;E-mail: wtl505@126.com
基金资助:
LIU Hui( ), LU Yang(
), LU Yang( ), YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa
), YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa
Received:2022-07-31
Published:2023-05-26
Online:2023-06-08
摘要:
Cd(镉)含量超标是影响苦荞(Fagopyrum tararicum)安全生产的主要重金属污染。为揭示外源添加硫(S)对苦荞耐Cd胁迫影响的分子机理,通过比较转录组学分析手段,分析根施100 mg/kg(NH4)2SO4(S)处理与5 mg/kg CdCl2(Cd)处理后苦荞叶片的差异表达基因及其功能。结果显示,共获得393 983 500条干净序列,鉴定到19 875个基因,其中差异表达基因共有1 760个。通过GO注释和KEGG pathway对这些差异表达基因进行分析表明,外源S添加增强谷胱甘肽还原酶途径,抑制氧化还原途径,并诱导水杨酸响应、脱落酸响应以及生长素响应途径应对Cd毒害。转录组数据鉴定到苦荞FtbHLH68受Cd+S处理特异诱导表达,模式植物拟南芥(Arabidopsis thaliana)同源基因突变体Atbhlh68表现出Cd+S处理缺陷表型,且Atbhlh68中叶绿素含量、净光合、GR活性和GSH活性变化均不响应Cd+S处理。研究结果揭示了苦荞响应Cd胁迫转录水平调控过程,阐明外源添加S增强植物抗Cd胁迫的分子机理,为外源S在苦荞抗Cd胁迫中的应用奠定理论基础并提供了作用靶点。
刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191.
LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat[J]. Biotechnology Bulletin, 2023, 39(5): 177-191.
| Gene | Primer-F(5'-3') | Primer-R(5'-3') |
|---|---|---|
| FtPinG0001636600.01 | CAAGCGCTTTCCAAGATCGG | GCACTCATGACTTCGTTCGC |
| FtPinG0007992900.01 | TGTTGTGTGTCCCACTCTGT | TTGGCTTGAGTCCCATGCTC |
| FtPinG0004475100.01 | GGCTTGATATTGCTGCGGTG | TTGCCAGCTCCTGCTTAGAC |
| FtPinG0002599400.01 | CACTTCTCCTCGCTAACGCT | CAATGGGTGAGTGGCGTAGT |
| FtPinG0005827500.01 | GTTCGGGAAGAAGTACGGGG | CCAGCTAACCCAACCTAGCC |
| FtPinG0000987900.01 | AAGTCCGTGGAGCAACTGAG | TGTGACTTCCTCCATGACGC |
| FtPinG0002633300.01 | AAGACCGCAGCGTACTGAAA | TCTCAAAGGCGCACAGCTTA |
| FtPinG0008441500.01 | GCCTTGGAGTGTCGACAAGA | TCCCTAACCCGAATGCACAC |
| FtPinG0001390600.01 | GCTGGCGAGGAATAATTGCG | GCGCAATCCTTGCACTTGAA |
| FtPinG0007214300.01 | AGGCATCACCAACATGGGAG | CTTGTGTGCTGAAAGTGCCC |
| FtPinG0002002200.01 | TCACGAGGCTGGTGTTTTGA | ATCTAGTGGCAACCGCGTAG |
| FtPinG0000414800.01 | GGGGAGAGCTCCATGTTGTG | TCCATGGCCGAATTGGTGAA |
表1 荧光定量PCR所用引物
Table 1 Primers for RT-qPCR
| Gene | Primer-F(5'-3') | Primer-R(5'-3') |
|---|---|---|
| FtPinG0001636600.01 | CAAGCGCTTTCCAAGATCGG | GCACTCATGACTTCGTTCGC |
| FtPinG0007992900.01 | TGTTGTGTGTCCCACTCTGT | TTGGCTTGAGTCCCATGCTC |
| FtPinG0004475100.01 | GGCTTGATATTGCTGCGGTG | TTGCCAGCTCCTGCTTAGAC |
| FtPinG0002599400.01 | CACTTCTCCTCGCTAACGCT | CAATGGGTGAGTGGCGTAGT |
| FtPinG0005827500.01 | GTTCGGGAAGAAGTACGGGG | CCAGCTAACCCAACCTAGCC |
| FtPinG0000987900.01 | AAGTCCGTGGAGCAACTGAG | TGTGACTTCCTCCATGACGC |
| FtPinG0002633300.01 | AAGACCGCAGCGTACTGAAA | TCTCAAAGGCGCACAGCTTA |
| FtPinG0008441500.01 | GCCTTGGAGTGTCGACAAGA | TCCCTAACCCGAATGCACAC |
| FtPinG0001390600.01 | GCTGGCGAGGAATAATTGCG | GCGCAATCCTTGCACTTGAA |
| FtPinG0007214300.01 | AGGCATCACCAACATGGGAG | CTTGTGTGCTGAAAGTGCCC |
| FtPinG0002002200.01 | TCACGAGGCTGGTGTTTTGA | ATCTAGTGGCAACCGCGTAG |
| FtPinG0000414800.01 | GGGGAGAGCTCCATGTTGTG | TCCATGGCCGAATTGGTGAA |
| 样品 Sample | 原始读取序列 Raw reads/bp | 干净读取序列总数 Total clean reads/bp | 总序列中质量值大于30的碱基数的比例 Clean Q30 bases rate/% | 比对到参考基因组序列比例 Mapping rate/% | 检测到表达的基因数目 Total gene |
|---|---|---|---|---|---|
| CK1 | 41 740 092 | 40 412 240 | 91.84 | 67.42 | 19 224 |
| CK2 | 46 423 990 | 45 073 886 | 92.23 | 68.31 | 19 362 |
| CK3 | 45 942 460 | 43 884 754 | 92.46 | 64.74 | 18 854 |
| Cd1 | 41 384 496 | 40 064 950 | 92.9 | 92.22 | 19 109 |
| Cd2 | 43 464 826 | 42 209 070 | 92.14 | 81.49 | 19 593 |
| Cd3 | 46 953 848 | 45 429 048 | 92.14 | 85.88 | 19 560 |
| Cd+S1 | 45 970 716 | 44 405 584 | 92.48 | 78.95 | 19 875 |
| Cd+S2 | 48 106 694 | 46 437 132 | 92.12 | 83.07 | 19 593 |
| Cd+S3 | 48 272 452 | 46 066 836 | 92.27 | 78.48 | 19 614 |
表2 样品分析结果统计表
Table 2 Statistical table of sample analysis results
| 样品 Sample | 原始读取序列 Raw reads/bp | 干净读取序列总数 Total clean reads/bp | 总序列中质量值大于30的碱基数的比例 Clean Q30 bases rate/% | 比对到参考基因组序列比例 Mapping rate/% | 检测到表达的基因数目 Total gene |
|---|---|---|---|---|---|
| CK1 | 41 740 092 | 40 412 240 | 91.84 | 67.42 | 19 224 |
| CK2 | 46 423 990 | 45 073 886 | 92.23 | 68.31 | 19 362 |
| CK3 | 45 942 460 | 43 884 754 | 92.46 | 64.74 | 18 854 |
| Cd1 | 41 384 496 | 40 064 950 | 92.9 | 92.22 | 19 109 |
| Cd2 | 43 464 826 | 42 209 070 | 92.14 | 81.49 | 19 593 |
| Cd3 | 46 953 848 | 45 429 048 | 92.14 | 85.88 | 19 560 |
| Cd+S1 | 45 970 716 | 44 405 584 | 92.48 | 78.95 | 19 875 |
| Cd+S2 | 48 106 694 | 46 437 132 | 92.12 | 83.07 | 19 593 |
| Cd+S3 | 48 272 452 | 46 066 836 | 92.27 | 78.48 | 19 614 |
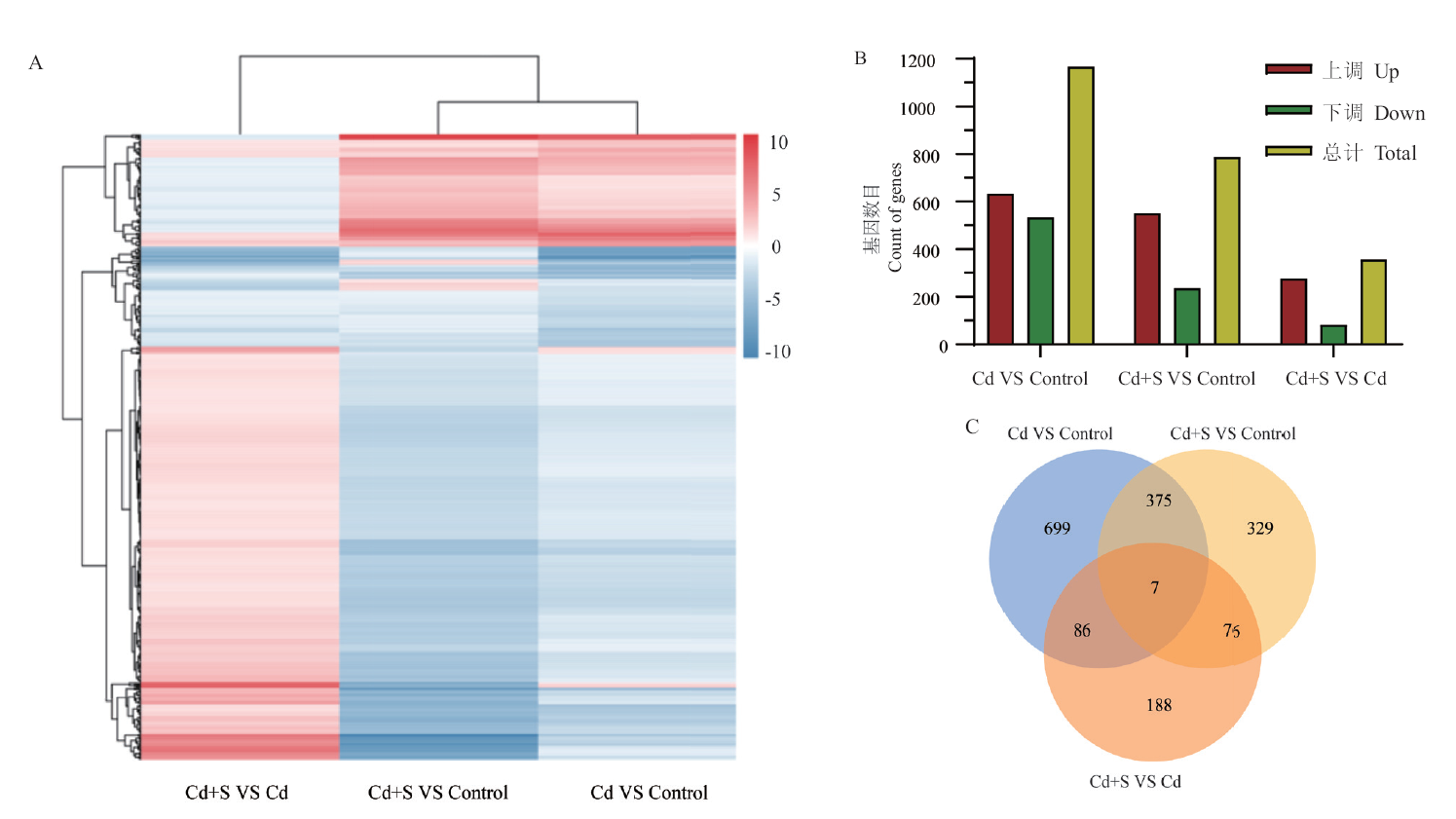
图1 黔苦4号叶片Cd处理和Cd+S处理的转录组分析结果 A:Cd处理和Cd+S处理转录组热图;B:Cd处理和Cd+S处理差异表达基因数量的统计;C:Cd处理和Cd+S处理差异表达基因韦恩图
Fig. 1 Transcriptome analysis of Cd treatment and Cd+S treatment in Qianku 4's leaves A: Transcriptome heat map in Cd treatment and Cd+S treatment. B: Statistics of the number of differentially expressed genes in Cd treatment and Cd+S treatment. C: Wayne diagram of differentially expressed genes under Cd treatment and Cd+S treatment
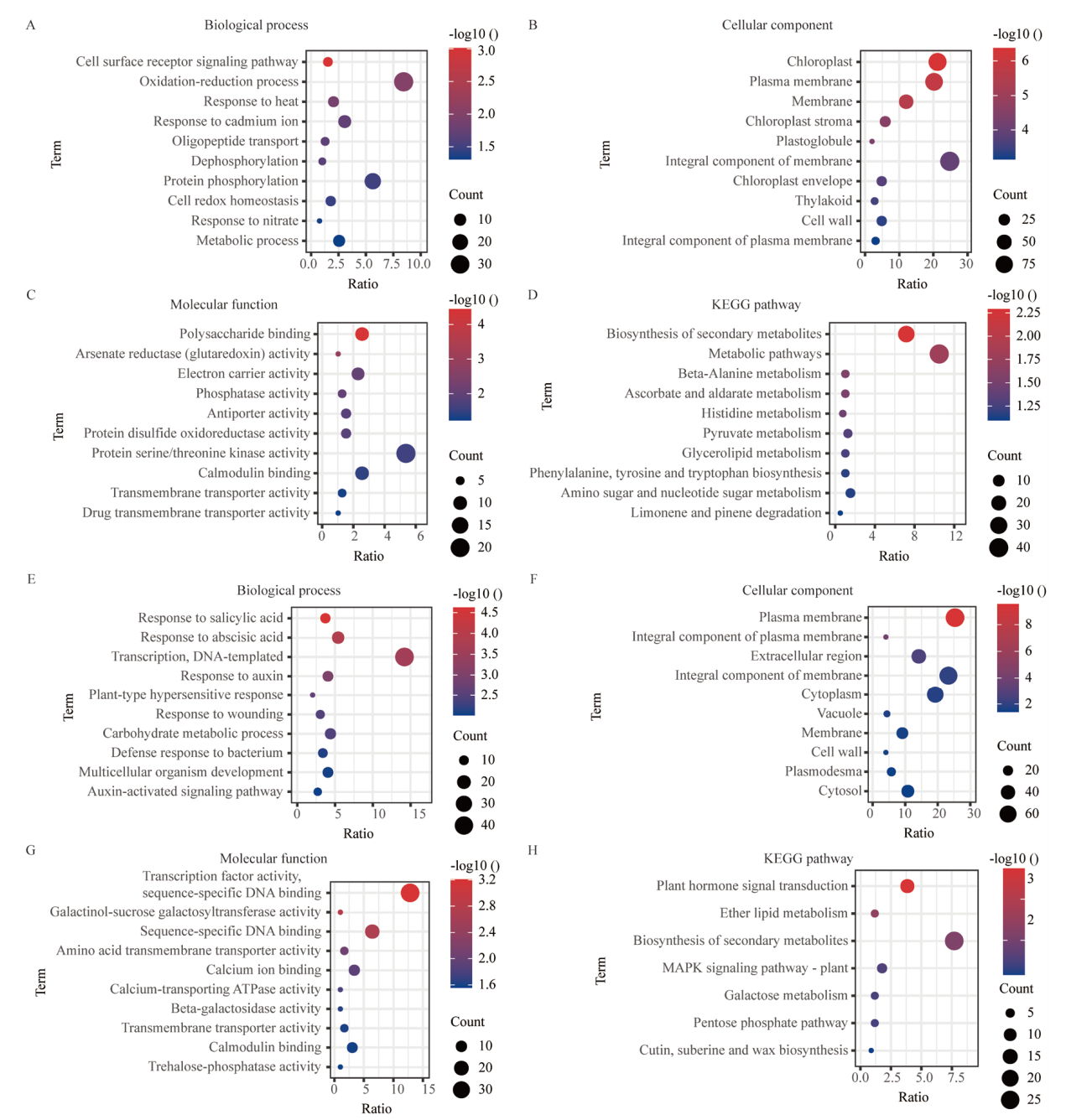
图2 Cd处理差异表达基因GO分析和KEGG分析 A-C:Cd处理上调基因GO分析;A:生物过程;B:细胞组分;C:分子功能;D:Cd处理上调基因KEGG分析;E-G:Cd处理下调基因GO分析;E:生物过程;F:细胞组分;G:分子功能;H:Cd处理下调基因KEGG分析
Fig. 2 GO analysis and KEGG analysis of differentially expressed genes under Cd treatment A-C: GO analysis of up-regulated genes under Cd treatment. A: Biological processes. B: Cellular components. C: Molecular function. D: KEGG analysis of up-regulated gene under Cd treatment. E-G: GO analysis of down-regulated genes under Cd treatment. E: Biological processes. F: Cell components. G: Molecular function. H: KEGG analysis of down-regulated gene under Cd treatment

图3 Cd+S处理差异表达基因GO分析和KEGG分析 A-C:Cd+S处理上调基因GO分析;A:生物过程;B:细胞组分;C:分子功能;D:Cd+S处理上调基因KEGG分析;E-G:Cd+S处理上调基因GO分析;E:生物过程;F:细胞组分;G:分子功能;H:Cd+S处理上调基因KEGG分析
Fig. 3 GO analysis and KEGG analysis of differentially expressed genes under Cd+S treatment A-C: GO analysis of up regulated genes under Cd+S treatment. A: Biological processes. B: Cellular components. C: Molecular function. D: KEGG analysis of up regulated gene under Cd+S treatment. E-G: GO analysis of down regulated genes under Cd+S treatment. E: Biological processes. F: Cell components. G: Molecular function. H: KEGG analysis of down regulated gene under Cd+S treatment
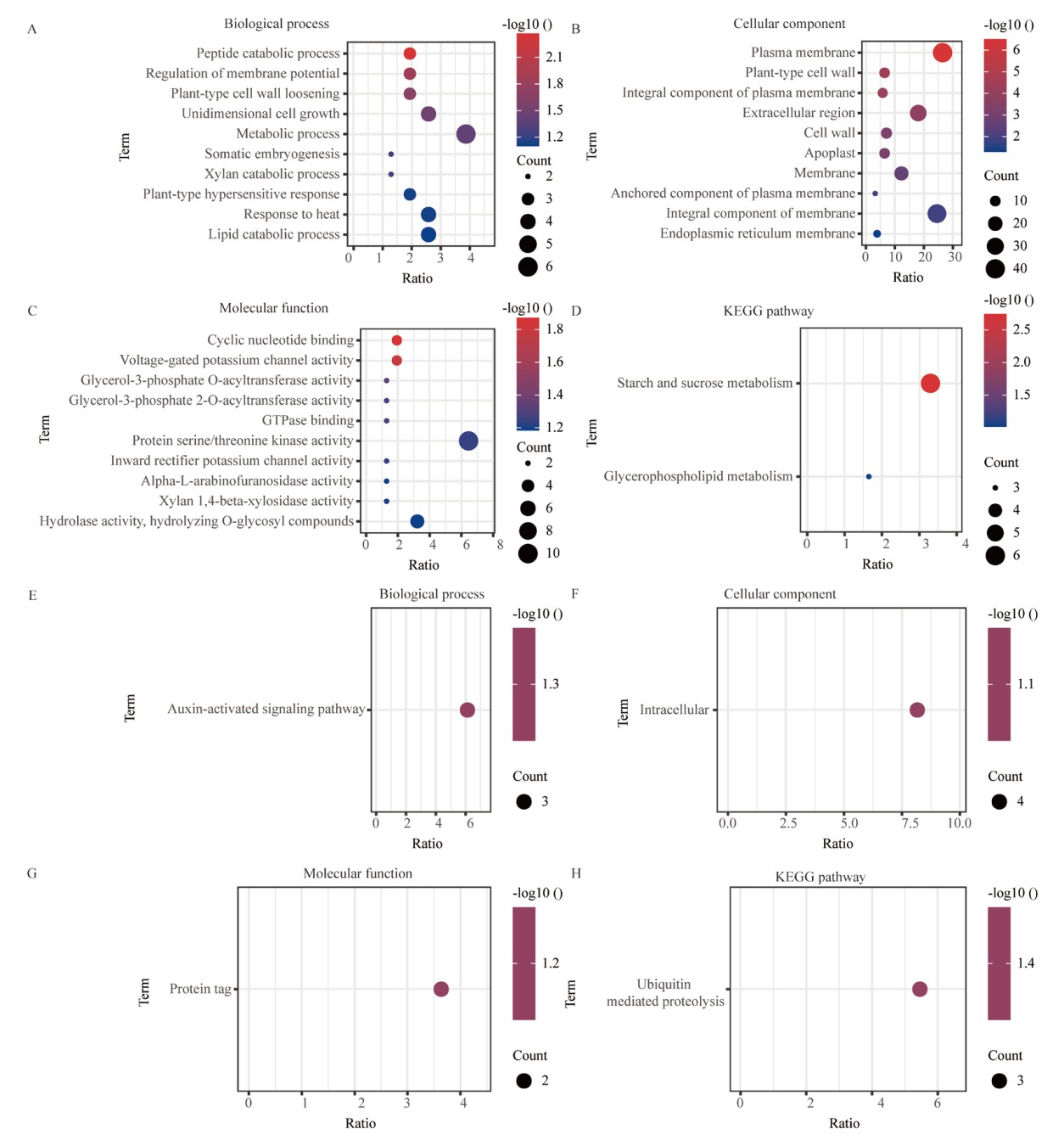
图4 Cd+S处理对比Cd处理差异表达基因GO分析和KEGG分析 A-C:Cd+S处理上调基因GO分析;A:生物过程;B:细胞组分;C:分子功能;D:Cd+S处理上调基因KEGG分析;E-G:Cd+S处理上调基因GO分析;E:生物过程;F:细胞组分;G:分子功能;H:Cd+S处理上调基因KEGG分析
Fig. 4 GO analysis and KEGG analysis of differentially expressed genes in CD+S treatment compared with CD treatment A-C: GO analysis of up-regulated genes under Cd+S treatment. A: Biological processes. B: Cellular components. C: Molecular function. D: KEGG analysis of up-regulated gene under Cd+S treatment. E-G: GO analysis of down-regulated genes under Cd+S treatment. E: Biological processes. F: Cell components. G: Molecular function. H: KEGG analysis of down-regulated gene under Cd+S treatment

图5 外源S添加影响Cd处理不同途径转录水平变化分析 A:Cd响应;B:氧化还原过程;C:谷胱甘肽还原酶;D:水杨酸响应;E:脱落酸响应;F:生长素响应;G: RT-qPCR对相关途径基因的验证
Fig. 5 Effects of exogenous S addition on transcriptional levels in different pathways of Cd treatment A: Response to cadmium ion. B: Oxidation-reduction process. C: Glutaredoxin activity. D: Response to salicylic acid. E: Response to abscisic acid. F: Response to auxin.G: Real-time quantitative PCR validation of several selected genes response to Cd stress and exogenous sulfur

图6 苦荞FtbHLH68基因功能验证 A:苦荞FtbHLH68基因表达检测;B-C:野生型Col-0和bhlh68突变体在Cd胁迫和Cd+S处理下的地上部分表型(B)以及叶片绿苗率(C);D-E:野生型Col-0和bhlh68突变体在Cd胁迫和Cd+S处理下的根长表型(D)以及统计结果(E);F-I:野生型Col-0和bhlh68突变体在Cd胁迫和Cd+S处理下叶绿素含量(F)、净光合(G)、GR活性(H)和GSH活性(I)考察。图中*表示在0.05水平差异显著
Fig. 6 Functional verification of tartary buckwheat FtbHLH68 gene A: Analysis of FtbHLH68 gene expression in tartary buckwheat. B-C: Phenotype (B) and green leaves rate (C) of wild-type Col-0 and bhlh68 mutants under Cd stress and Cd+S treatment. D-E: Root length phenotype (D) and statistical results (E) of wild-type Col-0 and bhlh68 mutants under Cd stress and Cd+S treatment. F-I: Chlorophyll content (F), net photosynthesis (G), GR activity (H) and GSH activity (I) of wild-type Col-0 and bhlh68 mutants under Cd stress and Cd+S treatment. * indicate significant difference at 0.05 level
| [1] |
Tang JB, Liu H, Lu Y, et al. Effect of exogenous sulfur on photosynthesis system of Fagopyrum tataricum seedlings under cadmium stress[J]. J Biobased Mat Bioenergy, 2021, 15(5): 705-712.
doi: 10.1166/jbmb.2021.2108 URL |
| [2] | Shanmugaraj BM, Malla A, Ramalingam S. Cadmium stress and toxicity in plants: an overview[M]// Cadmium toxicity and tolerance in plants. Amsterdam: Elsevier, 2019: 1-17. |
| [3] | Suhani I, Sahab S, Srivastava V, et al. Impact of cadmium pollution on food safety and human health[J]. Curr Opin Toxicol, 2021, 27: 1-7. |
| [4] |
Hussain B, Ashraf MN, Shafeeq-Ur-Rahman, et al. Cadmium stress in paddy fields: effects of soil conditions and remediation strategies[J]. Sci Total Environ, 2021, 754: 142188.
doi: 10.1016/j.scitotenv.2020.142188 URL |
| [5] | Dowidar SMA, Abo-Hamad SA, Mohsen AA, et al. Bioremediation of copper stressed Trigonella foenum graecum[J]. Journal of Stress Physiology & Biochemistry, 2013, 9(4): 5-24. |
| [6] |
Cho UH, Seo NH. Oxidative stress in Arabidopsis thaliana exposed to cadmium is due to hydrogen peroxide accumulation[J]. Plant Sci, 2005, 168(1): 113-120.
doi: 10.1016/j.plantsci.2004.07.021 URL |
| [7] |
Redjala T, Zelko I, Sterckeman T, et al. Relationship between root structure and root cadmium uptake in maize[J]. Environ Exp Bot, 2011, 71(2): 241-248.
doi: 10.1016/j.envexpbot.2010.12.010 URL |
| [8] |
El Rasafi T, Oukarroum A, Haddioui A, et al. Cadmium stress in plants: a critical review of the effects, mechanisms, and tolerance strategies[J]. Crit Rev Environ Sci Technol, 2022, 52(5): 675-726.
doi: 10.1080/10643389.2020.1835435 URL |
| [9] |
Gallego SM, Pena LB, Barcia RA, et al. Unravelling cadmium toxicity and tolerance in plants: insight into regulatory mechanisms[J]. Environ Exp Bot, 2012, 83: 33-46.
doi: 10.1016/j.envexpbot.2012.04.006 URL |
| [10] |
Wu ZC, Zhao XH, Sun XC, et al. Xylem transport and gene expression play decisive roles in cadmium accumulation in shoots of two oilseed rape cultivars(Brassica napus)[J]. Chemosphere, 2015, 119: 1217-1223.
doi: 10.1016/j.chemosphere.2014.09.099 URL |
| [11] |
Herbette S, Taconnat L, Hugouvieux V, et al. Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots[J]. Biochimie, 2006, 88(11): 1751-1765.
pmid: 16797112 |
| [12] |
He M, He CQ, Ding NZ. Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance[J]. Front Plant Sci, 2018, 9: 1771.
doi: 10.3389/fpls.2018.01771 pmid: 30581446 |
| [13] | Bali AS, Sidhu GPS, Kumar V, et al. Mitigating cadmium toxicity in plants by phytohormones[M]// Cadmium toxicity and tolerance in plants. Amsterdam: Elsevier, 2019: 375-396. |
| [14] | Hameed A, Rasool S, Azooz MM, et al. Heavy metal stress: plant responses and signaling[M]// Plant metal interaction. Amsterdam: Elsevier, 2015: 557-583. |
| [15] |
Kumar A, Prasad MNV. Plant-lead interactions: transport, toxicity, tolerance, and detoxification mechanisms[J]. Ecotoxicol Environ Saf, 2018, 166: 401-418.
doi: 10.1016/j.ecoenv.2018.09.113 URL |
| [16] |
Mahmud JA, Hasanuzzaman M, Nahar K, et al. Insights into citric acid-induced cadmium tolerance and phytoremediation in Brassica juncea L.: Coordinated functions of metal chelation, antioxidant defense and glyoxalase systems[J]. Ecotoxicol Environ Saf, 2018, 147: 990-1001.
doi: 10.1016/j.ecoenv.2017.09.045 URL |
| [17] |
Takahashi H, Kopriva S, Giordano M, et al. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes[J]. Annu Rev Plant Biol, 2011, 62: 157-184.
doi: 10.1146/annurev-arplant-042110-103921 pmid: 21370978 |
| [18] |
Lunde C, Zygadlo A, Simonsen HT, et al. Sulfur starvation in rice: the effect on photosynthesis, carbohydrate metabolism, and oxidative stress protective pathways[J]. Physiol Plant, 2008, 134(3): 508-521.
doi: 10.1111/j.1399-3054.2008.01159.x pmid: 18785901 |
| [19] |
Gill SS, Tuteja N. Cadmium stress tolerance in crop plants: probing the role of sulfur[J]. Plant Signal Behav, 2011, 6(2): 215-222.
doi: 10.4161/psb.6.2.14880 pmid: 21330784 |
| [20] |
Zaid A, Bhat JA, Wani SH, et al. Role of nitrogen and sulfur in mitigating cadmium induced metabolism alterations in plants[J]. J Plant Sci Res, 2019, 35(1): 121-141.
doi: 10.32381/JPSR URL |
| [21] |
Mendoza-Cózatl D, Loza-Tavera H, Hernández-Navarro A, et al. Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants[J]. FEMS Microbiol Rev, 2005, 29(4): 653-671.
pmid: 16102596 |
| [22] |
Fan JL, Hu ZY, Ziadi N, et al. Excessive sulfur supply reduces cadmium accumulation in brown rice(Oryza sativa L.)[J]. Environ Pollut, 2010, 158(2): 409-415.
doi: 10.1016/j.envpol.2009.08.042 URL |
| [23] |
Zhang DX, Du GH, Chen D, et al. Effect of elemental sulfur and gypsum application on the bioavailability and redistribution of cadmium during rice growth[J]. Sci Total Environ, 2019, 657: 1460-1467.
doi: 10.1016/j.scitotenv.2018.12.057 URL |
| [24] |
Cao ZZ, Qin ML, Lin XY, et al. Sulfur supply reduces cadmium uptake and translocation in rice grains(Oryza sativa L.) by enhancing iron plaque formation, cadmium chelation and vacuolar sequestration[J]. Environ Pollut, 2018, 238: 76-84.
doi: 10.1016/j.envpol.2018.02.083 URL |
| [25] |
Adhikari S, Ghosh S, Azahar I, et al. Sulfate improves cadmium tolerance by limiting cadmium accumulation, modulation of sulfur metabolism and antioxidant defense system in maize[J]. Environ Exp Bot, 2018, 153: 143-162.
doi: 10.1016/j.envexpbot.2018.05.008 URL |
| [26] | 余霜, 李光, 陈庆富. 贵州省苦荞产业发展的SWOT-PEST分析[J]. 南方农业, 2016, 10(34): 27-29. |
| Yu S, Li G, Chen QF. SWOT-PEST analysis of Tartary buckwheat industry development in Guizhou Province[J]. South China Agric, 2016, 10(34): 27-29. | |
| [27] | 宋春然, 何锦林, 谭红, 等. 贵州省农业土壤重金属污染的初步评价[J]. 贵州农业科学, 2005, 33(2): 13-16. |
| Song CR, He JL, Tan H, et al. Primary appraisal for heavy metals pollution in farm soils of Guizhou Province[J]. Guizhou Agric Sci, 2005, 33(2): 13-16. | |
| [28] | 毛旭, 付天岭, 何腾兵, 等. 苦荞低镉积累品种筛选及富集转运特征分析[J]. 地球与环境, 2022, 50(1): 103-109. |
| Mao X, Fu TL, He TB, et al. Screening of low cadmium accumulation cultivars of Tartary buckwheat and analysis of the characteristics of bioconcentration and transportation[J]. Earth Environ, 2022, 50(1): 103-109. | |
| [29] | Ye XL, Liu CY, Yan HL, et al. Genome-wide identification and transcriptome analysis of the heavy metal-associated(HMA)gene family in Tartary buckwheat and their regulatory roles under cadmium stress[J]. Genes, 2022, 847: 146884. |
| [30] |
Lu Y, Wang QF, Li J, et al. Effects of exogenous sulfur on alleviating cadmium stress in Tartary buckwheat[J]. Sci Rep, 2019, 9(1): 7397.
doi: 10.1038/s41598-019-43901-4 pmid: 31089197 |
| [31] |
Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation[J]. Nat Biotechnol, 2010, 28(5): 511-515.
doi: 10.1038/nbt.1621 pmid: 20436464 |
| [32] |
Yan LH, Wei SW, Wu YR, et al. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system[J]. Mol Plant, 2015, 8(12): 1820-1823.
doi: 10.1016/j.molp.2015.10.004 URL |
| [33] |
Wu XL, Yuan J, Luo AX, et al. Drought stress and re-watering increase secondary metabolites and enzyme activity in Dendrobium moniliforme[J]. Ind Crops Prod, 2016, 94: 385-393.
doi: 10.1016/j.indcrop.2016.08.041 URL |
| [34] |
Nemmiche S. Oxidative signaling response to cadmium exposure[J]. Toxicol Sci, 2017, 156(1): 4-10.
doi: 10.1093/toxsci/kfw222 pmid: 27803385 |
| [35] |
Joseph P. Mechanisms of cadmium carcinogenesis[J]. Toxicol Appl Pharmacol, 2009, 238(3): 272-279.
doi: 10.1016/j.taap.2009.01.011 URL |
| [36] |
Benavides MP, Gallego SM, Tomaro ML. Cadmium toxicity in plants[J]. Braz J Plant Physiol, 2005, 17(1): 21-34.
doi: 10.1590/S1677-04202005000100003 URL |
| [37] |
Haider FU, Cai LQ, Coulter JA, et al. Cadmium toxicity in plants: impacts and remediation strategies[J]. Ecotoxicol Environ Saf, 2021, 211: 111887.
doi: 10.1016/j.ecoenv.2020.111887 URL |
| [38] |
Iannelli MA, Pietrini F, Fiore L, et al. Antioxidant response to cadmium in Phragmites australis plants[J]. Plant Physiol Biochem, 2002, 40(11): 977-982.
doi: 10.1016/S0981-9428(02)01455-9 URL |
| [39] |
Chien HF, Lin CC, Wang JW, et al. Changes in ammonium ion content and glutamine synthetase activity in rice leaves caused by excess cadmium are a consequence of oxidative damage[J]. Plant Growth Regul, 2002, 36: 41-47.
doi: 10.1023/A:1014742014171 URL |
| [40] |
Olmos E, Martínez-Solano JR, Piqueras A, et al. Early steps in the oxidative burst induced by cadmium in cultured tobacco cells (BY-2 line)[J]. J Exp Bot, 2003, 54(381): 291-301.
doi: 10.1093/jxb/erg028 pmid: 12493856 |
| [41] |
Hendrix S, Jozefczak M, Wójcik M, et al. Glutathione: a key player in metal chelation, nutrient homeostasis, cell cycle regulation and the DNA damage response in cadmium-exposed Arabidopsis thaliana[J]. Plant Physiol Biochem, 2020, 154: 498-507.
doi: 10.1016/j.plaphy.2020.06.006 URL |
| [42] |
Noctor G, Mhamdi A, Chaouch S, et al. Glutathione in plants: an integrated overview[J]. Plant Cell Environ, 2012, 35(2): 454-484.
doi: 10.1111/j.1365-3040.2011.02400.x URL |
| [43] |
Popova LP, Maslenkova LT, Yordanova RY, et al. Exogenous treatment with salicylic acid attenuates cadmium toxicity in pea seedlings[J]. Plant Physiol Biochem, 2009, 47(3): 224-231.
doi: 10.1016/j.plaphy.2008.11.007 URL |
| [44] |
Liu ZP, Ding YF, Wang FJ, et al. Role of salicylic acid in resistance to cadmium stress in plants[J]. Plant Cell Rep, 2016, 35(4): 719-731.
doi: 10.1007/s00299-015-1925-3 pmid: 26849671 |
| [45] |
Chen K, Li GJ, Bressan RA, et al. Abscisic acid dynamics, signaling, and functions in plants[J]. J Integr Plant Biol, 2020, 62(1): 25-54.
doi: 10.1111/jipb.12899 |
| [46] | 韩超, 申海玉, 叶嘉, 等. 外源脱落酸对小麦幼苗抗镉胁迫能力的影响[J]. 西北植物学报, 2012, 32(4): 745-750. |
| Han C, Shen HY, Ye J, et al. Effect of exogenous abscisic acid on tolerance of wheat seedlings to cadmium stress[J]. Acta Bot Boreali Occidentalia Sin, 2012, 32(4): 745-750. | |
| [47] |
Shen GM, Niu JK, Deng ZX. Abscisic acid treatment alleviates cadmium toxicity in purple flowering stalk(Brassica campestris L. ssp. chinensis var. purpurea Hort.)seedlings[J]. Plant Physiol Biochem, 2017, 118: 471-478.
doi: 10.1016/j.plaphy.2017.07.018 URL |
| [48] |
Hsu YT, Kao CH. Role of abscisic acid in cadmium tolerance of rice(Oryza sativa L.) seedlings[J]. Plant Cell Environ, 2003, 26(6): 867-874.
doi: 10.1046/j.1365-3040.2003.01018.x URL |
| [49] |
Vanneste S, Friml J. Auxin: a trigger for change in plant development[J]. Cell, 2009, 136(6): 1005-1016.
doi: 10.1016/j.cell.2009.03.001 pmid: 19303845 |
| [50] |
Yuan HM, Huang X. Inhibition of root meristem growth by cadmium involves nitric oxide-mediated repression of auxin accumulation and signalling in Arabidopsis[J]. Plant Cell Environ, 2016, 39(1): 120-135.
doi: 10.1111/pce.12597 URL |
| [51] |
Luo Y, Wei YW, Sun SG, et al. Selenium modulates the level of auxin to alleviate the toxicity of cadmium in tobacco[J]. Int J Mol Sci, 2019, 20(15): 3772.
doi: 10.3390/ijms20153772 URL |
| [52] |
Zhou C, Zhu L, Ma ZY, et al. Bacillus amyloliquefaciens SAY09 increases cadmium resistance in plants by activation of auxin-mediated signaling pathways[J]. Genes, 2017, 8(7): 173.
doi: 10.3390/genes8070173 URL |
| [53] | Zhu XF, Wang ZW, Dong F, et al. Exogenous auxin alleviates cadmium toxicity in Arabidopsis thaliana by stimulating synthesis of hemicellulose 1 and increasing the cadmium fixation capacity of root cell walls[J]. J Hazard Mater, 2013, 263 Pt 2: 398-403. |
| [54] |
Hao YQ, Zong XM, Ren P, et al. Basic helix-loop-helix(bHLH)transcription factors regulate a wide range of functions in Arabidopsis[J]. Int J Mol Sci, 2021, 22(13): 7152.
doi: 10.3390/ijms22137152 URL |
| [55] |
Stroiński A, Chadzinikolau T, Giżewska K, et al. ABA or cadmium induced phytochelatin synthesis in potato tubers[J]. Biol Plant, 2010, 54(1): 117-120.
doi: 10.1007/s10535-010-0017-z URL |
| [56] |
Li CJ, Yan CX, Sun QX, et al. The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut[J]. BMC Plant Biol, 2021, 21(1): 540.
doi: 10.1186/s12870-021-03318-6 pmid: 34784902 |
| [57] | Liang YF, Ma F, Li BY, et al. A bHLH transcription factor SlbHLH96 promotes drought tolerance in tomato[J]. Hortic Res, 2022. https://doi.org/10.1093/hr/uhac198. |
| [58] |
Hussain Q, Asim M, Zhang R, et al. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress[J]. Biomolecules, 2021, 11(8): 1159.
doi: 10.3390/biom11081159 URL |
| [59] |
Le HR, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana[J]. Physiol Plant, 2017, 160(3): 312-327.
doi: 10.1111/ppl.2017.160.issue-3 URL |
| [60] |
Lu QY, Chen SM, Li YY, et al. Exogenous abscisic acid(ABA)promotes cadmium(Cd)accumulation in Sedum alfredii Hance by regulating the expression of Cd stress response genes[J]. Environ Sci Pollut Res Int, 2020, 27(8): 8719-8731.
doi: 10.1007/s11356-019-07512-w |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [4] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [5] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [6] | 孙言秋, 谢采芸, 汤岳琴. 耐高温酿酒酵母的构建与高温耐受机制解析[J]. 生物技术通报, 2023, 39(11): 226-237. |
| [7] | 姜南, 石杨, 赵志慧, 李斌, 赵熠辉, 杨俊彪, 闫家铭, 靳雨璠, 陈稷, 黄进. 镉胁迫下水稻OsPT1的表达及功能分析[J]. 生物技术通报, 2023, 39(1): 166-174. |
| [8] | 关志秀, 汪燕, 梁成刚, 韦春玉, 黄娟, 陈庆富. 苦荞FtCBL基因的鉴定及对干旱与高钙胁迫的响应[J]. 生物技术通报, 2022, 38(8): 101-109. |
| [9] | 呼艳姣, 陈美凤, 强瑀, 李海燕, 刘静, 秦樊鑫. 镉胁迫下锌硒交互作用对水稻镉毒害的缓解机制[J]. 生物技术通报, 2022, 38(4): 143-152. |
| [10] | 祖国蔷, 胡哲, 王琪, 李光哲, 郝林. Burkholderia sp. GD17对水稻幼苗镉耐受的调节[J]. 生物技术通报, 2022, 38(4): 153-162. |
| [11] | 胡琪, 侯玉翔, 李璿, 李梅兰. 普通白菜CYP79B2同源基因的克隆与表达[J]. 生物技术通报, 2022, 38(12): 168-174. |
| [12] | 钱静洁, 林苏梦, 张冬平, 高勇. 光敏色素互作因子参与生长素调控的植物生长发育[J]. 生物技术通报, 2022, 38(10): 29-33. |
| [13] | 唐嘉城, 梁毅珉, 马葭思, 彭桂香, 谭志远. 百香果内生细菌多样性及促生长特性[J]. 生物技术通报, 2022, 38(1): 86-97. |
| [14] | 杨馥榕, 王晓红, 肖琪, 方娟, 李立华. 木槿品种对镉胁迫的生理响应及耐镉能力评价[J]. 生物技术通报, 2022, 38(1): 98-107. |
| [15] | 孙小倩, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子FtMYBF的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2021, 37(3): 10-17. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||