








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 10-23.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0985
收稿日期:2022-08-13
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
于洪巍,男,教授,研究方向:蛋白质工程、代谢工程;E-mail: yuhongwei@zju.edu.cn;作者简介:张岩峰,男,硕士研究生,研究方向:蛋白质工程;E-mail: 22028069@zju.edu.cn
基金资助:
ZHANG Yan-feng( ), YE Li-dan(
), YE Li-dan( ), YU Hong-wei(
), YU Hong-wei( )
)
Received:2022-08-13
Published:2023-04-26
Online:2023-05-16
摘要:
细胞色素P450酶(CYPs或P450s)可将O2的一个原子插入有机底物同时将另一个原子还原为水,广泛参与各种合成代谢和分解代谢过程,所以一直以来都是生物技术领域关注的焦点。在催化循环底物的氧化依赖于氧化还原伴侣向血红素铁传递电子,因此电子转移是P450s催化过程中的限速步骤。利用不同方法优化蛋白质-蛋白质相互作用以提高P450系统的电子转移效率,被称为“氧化还原伴侣工程”,是目前工程化P450s的重要手段之一,并取得了卓有成效的进展。本文将着重介绍关于氧化还原伴侣组分替换组装、P450酶与氧化还原伴侣融合及P450酶与氧化还原伴侣作用界面修饰等方面的进展,期望为未来该方面的工作提供一定的指导作用。
张岩峰, 叶丽丹, 于洪巍. 氧化还原伴侣工程:P450低效问题的解决方案之一[J]. 生物技术通报, 2023, 39(4): 10-23.
ZHANG Yan-feng, YE Li-dan, YU Hong-wei. Redox Partner Engineering: A Solution to the Low Catalytic Efficiency of P450s[J]. Biotechnology Bulletin, 2023, 39(4): 10-23.
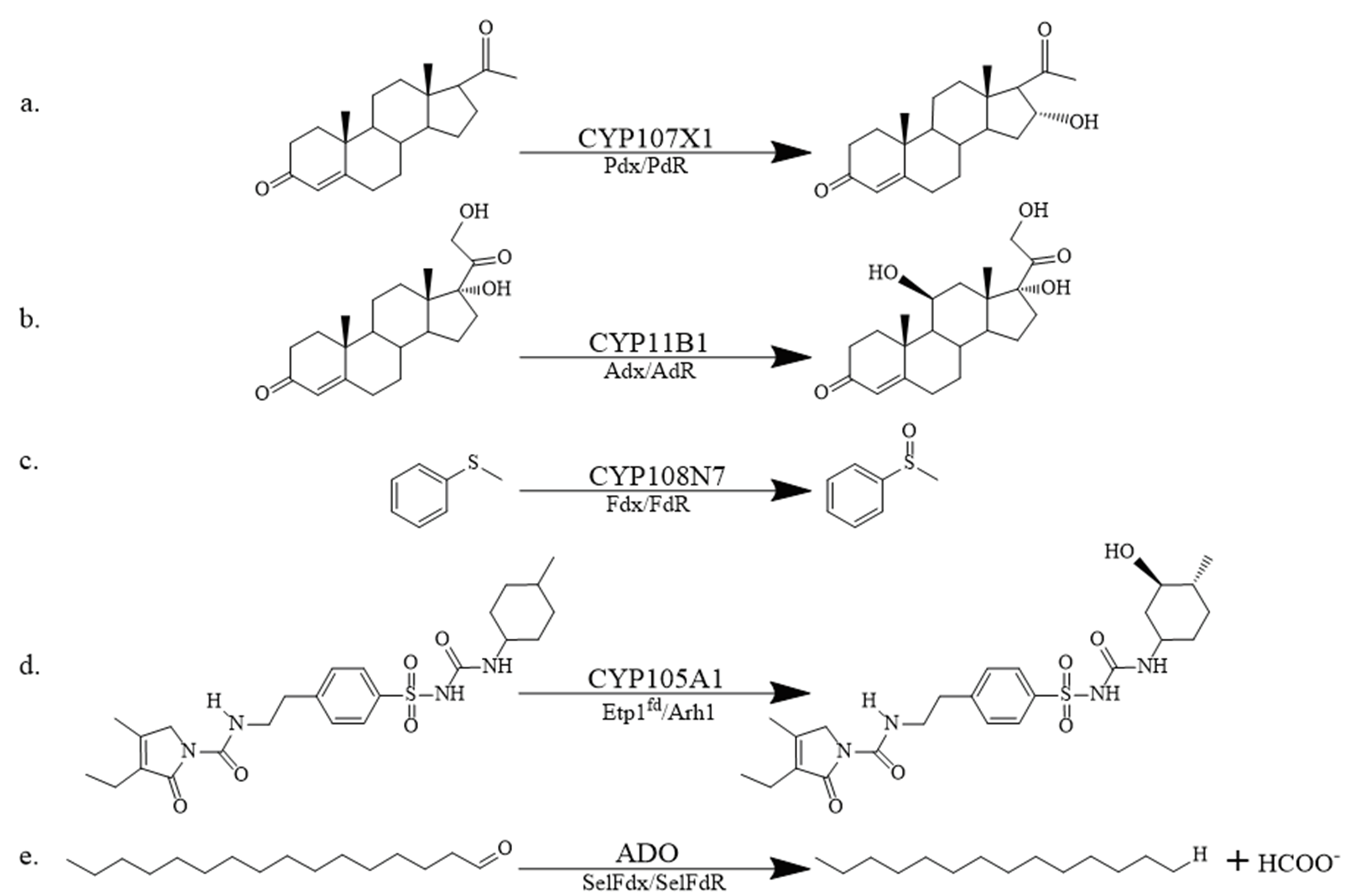
图2 经典RPs支持的部分氧化还原反应 a:孕酮的16α-羟基化;b: 11-脱氧皮质酮的11β-羟基化;c:苯甲硫醚的硫氧化;d:磺酰脲类药物格列美脲的3-环己基羟基化;e:十六醛的脱甲酰化
Fig. 2 Examples of partial redox reactions supported by classical RPs a: 16α-hydroxylation of progesterone; b: 11β-hydroxylation of 11-deoxycorticosterone; c: sulfur oxidation of thioanisole; d: 3-cyclohexyl-hydroxylation of the sulfonylurea drug glimepiride; e: deformylation of n-hexadecanal
| RPs | P450酶 Enzyme P450 | 底物Substrate | 组装结果Assembly results | 实例Example |
|---|---|---|---|---|
| P. putida Pdx/PdR | StreptomycesCYP154C3s | 孕酮 | 支持对10种类固醇底物的反应活性 | [ |
| S. avermitilisCYP107X1 | 孕酮 | 支持孕酮的16α-羟基化 | [ | |
| E. coli Fld/Fpr | 牛CYP17A1 | 孕酮 | 支持孕酮的17α-羟基化 | [ |
| S. cellulosumCYP264A1 B. megateriumCYP106A2 | 4-甲基-3-苯基香豆素,11-脱氧皮质酮 | Adx/Fpr比Adx/AdR 对CYP264A1和CYP106A2的反应初始速率分别提升52%和33% | [ | |
| 菠菜Fdx/FdR | R.wratislaviensis CYP108N7 | 苯甲硫醚 | 比其他RPs的转化率高5倍以上 | [ |
| 哺乳动物Adx/AdR | B. megateriumCYP106A2 | 11-脱氧皮质酮 | 支持转化速率达到1 mmol/(L·d) | [ |
| 人CYP11B1 | 11-脱氧皮质醇 | 支持皮质醇产量达0.84 g/(L·d) | [ | |
| S. pombe Etp1fd/Arh1 | S. griseolusCYP105A1 | 磺酰脲类 | 支持3-环己基羟基化活性 | [ |
| 牛CYP21A2 | Madrane | 比同源CPR的时空产率高1.8倍 | [ | |
| S. elongatusSelFdx1499/SelFdR0978 | S. elongatusADO(非P450酶) | 正十五烷 | 比菠菜Fdx/FdD产率高26.6% | [ |
| S. benihana CYP-sb21 | 环孢菌素 | 比其他RPs支持的转化率高1.4倍以上 | [ | |
| B. megaterium Fdx2 /BmCPR | B. megateriumCYP106A1 | 11-酮基-β-乳香酸 | Fdx2/Arh1支持最高的转化率(79.6±6.9)% | [ |
| 牛CYP21A2 | 孕酮 | BmCPR比同源CPR催化效率高1.7倍 | [ | |
| S. coelicolor Fdx4/ FDR1 | S. coelicolorCYP105D5 | 脂肪酸 | 比其他同源和异源RPs催化效率均高 | [ |
| S. Cellulosum Fdxs/FdR_B | S. CellulosumCYP260A1 | 诺卡酮 | Fdx2/FdR_B比其他同源RPs转化率均高 | [ |
| S. CellulosumCYP109D1 | 脂肪酸 | Fdx8/FdR_B比Fdx2/FdR_B有效,但产物生成效率比牛Adx/AdR低3倍 | [ | |
| Synechocystis SynFdx/C. reinhardtii FNR | S. cellulosumP450 EpoK | 埃博霉素D | SynFdx/ FNR比其他RPs组合转化率高8-11倍 | [ |
| S. cerevisiae CPR | Aspergillus oryzaeCYPs | 7-乙氧基香豆素 | 支持多个P450酶的催化活性 | [ |
| T. cuspidata CPR | T. cuspidata 10β-羟化酶 | 紫杉烯-5α-乙酸酯 | 比酵母CPR羟化活性提升7倍 | [ |
| C. apicola CPR | B. subtilisCYP109B1,T. fusca CYP154E1 | 肉豆蔻酸,香叶醇 | 支持CYP109B1和CYP154E1的羟化活性 | [ |
表1 RPs替换组装实例
Table 1 Replacement and assembly examples of RPs
| RPs | P450酶 Enzyme P450 | 底物Substrate | 组装结果Assembly results | 实例Example |
|---|---|---|---|---|
| P. putida Pdx/PdR | StreptomycesCYP154C3s | 孕酮 | 支持对10种类固醇底物的反应活性 | [ |
| S. avermitilisCYP107X1 | 孕酮 | 支持孕酮的16α-羟基化 | [ | |
| E. coli Fld/Fpr | 牛CYP17A1 | 孕酮 | 支持孕酮的17α-羟基化 | [ |
| S. cellulosumCYP264A1 B. megateriumCYP106A2 | 4-甲基-3-苯基香豆素,11-脱氧皮质酮 | Adx/Fpr比Adx/AdR 对CYP264A1和CYP106A2的反应初始速率分别提升52%和33% | [ | |
| 菠菜Fdx/FdR | R.wratislaviensis CYP108N7 | 苯甲硫醚 | 比其他RPs的转化率高5倍以上 | [ |
| 哺乳动物Adx/AdR | B. megateriumCYP106A2 | 11-脱氧皮质酮 | 支持转化速率达到1 mmol/(L·d) | [ |
| 人CYP11B1 | 11-脱氧皮质醇 | 支持皮质醇产量达0.84 g/(L·d) | [ | |
| S. pombe Etp1fd/Arh1 | S. griseolusCYP105A1 | 磺酰脲类 | 支持3-环己基羟基化活性 | [ |
| 牛CYP21A2 | Madrane | 比同源CPR的时空产率高1.8倍 | [ | |
| S. elongatusSelFdx1499/SelFdR0978 | S. elongatusADO(非P450酶) | 正十五烷 | 比菠菜Fdx/FdD产率高26.6% | [ |
| S. benihana CYP-sb21 | 环孢菌素 | 比其他RPs支持的转化率高1.4倍以上 | [ | |
| B. megaterium Fdx2 /BmCPR | B. megateriumCYP106A1 | 11-酮基-β-乳香酸 | Fdx2/Arh1支持最高的转化率(79.6±6.9)% | [ |
| 牛CYP21A2 | 孕酮 | BmCPR比同源CPR催化效率高1.7倍 | [ | |
| S. coelicolor Fdx4/ FDR1 | S. coelicolorCYP105D5 | 脂肪酸 | 比其他同源和异源RPs催化效率均高 | [ |
| S. Cellulosum Fdxs/FdR_B | S. CellulosumCYP260A1 | 诺卡酮 | Fdx2/FdR_B比其他同源RPs转化率均高 | [ |
| S. CellulosumCYP109D1 | 脂肪酸 | Fdx8/FdR_B比Fdx2/FdR_B有效,但产物生成效率比牛Adx/AdR低3倍 | [ | |
| Synechocystis SynFdx/C. reinhardtii FNR | S. cellulosumP450 EpoK | 埃博霉素D | SynFdx/ FNR比其他RPs组合转化率高8-11倍 | [ |
| S. cerevisiae CPR | Aspergillus oryzaeCYPs | 7-乙氧基香豆素 | 支持多个P450酶的催化活性 | [ |
| T. cuspidata CPR | T. cuspidata 10β-羟化酶 | 紫杉烯-5α-乙酸酯 | 比酵母CPR羟化活性提升7倍 | [ |
| C. apicola CPR | B. subtilisCYP109B1,T. fusca CYP154E1 | 肉豆蔻酸,香叶醇 | 支持CYP109B1和CYP154E1的羟化活性 | [ |
| RPs | P450酶Enzyme P450 | 底物Substrate | 融合结果Fusion results | 实例Example |
|---|---|---|---|---|
| B. megateriumP450 BM3还原酶(BMR) | 哺乳动物肝脏CYPs | 多种脂肪酸和药物 | 支持多个P450酶的催化活性,真核P450酶的溶解度明显提升 | [ |
| A. OryzaeCYP57B3 | 大豆苷元 | 支持6-OH大豆苷元最大产量为9.1 mg/L | [ | |
| JeotgalicoccusP450 OleTJE | 饱和脂肪酸 | 比Pdx/PdR的周转率高8.9倍 | [ | |
| B. subtilisCYP102A3还原酶 | B. megateriumP450 BM3血红素域 | 12-对硝基苯基十二烷酸 | 比天然酶的热稳定性明显提升 | [ |
| RhodococcusCYP116B2还原酶(RhFRED) | S. venezuelaeP450 PikC | 12元环大环内酯YC-17和14元环大环内酯那博霉素 | 比菠菜Fdx/FdR的催化活性提高约4倍 | [ |
| StreptomycesP450 TamI | 去氧糖胺糖苷修饰的碳环 | 替代菠菜Fdx/FdR实现了制备级规模的C10羟化反应 | [ | |
| A. orientalisP450Prava | 紧缩素 | 支持在产黄青霉中产生超过6 g/L的普伐他汀 | [ | |
| M. griseorubidaP450 MycG | 麦新米星 | 支持P450 MycG的常规羟化反应 | [ | |
| R. ruberCYP116B3还原酶(PFOR) | M. aquaeoleiCYP153AM.aq | 十二烷酸 | 比融合RhFRED的反应初始速率高3倍 | [ |
| T. thermophilesCYP116B46还原酶 | R. coprophilusTC-2 P450tol | 甲苯 | 比融合RhFRED的半衰期和活性分别提高20倍和2倍以上 | [ |
| 大鼠CPR | 大鼠CYP1A1 | 7-乙氧基香豆素 | 融合酶比无CPR的CYP1A1催化活性高4倍 | [ |
| ArabidopsisCPR | P. ginseng P450 PPDS | 达马烯二醇 | 比共表达CPR的催化活性高约4.5倍 | [ |
| T. cuspidataCPR | T. cuspidataCYP725A4 | 紫衫二烯 | 比共表达CPR的催化活性显著降低 | [ |
| H. tuberosusCPR | H. tuberosusCYP76B1 | 苯脲类除草剂 | 比只存在CYP76B1的除草剂耐受性降低一半以上 | [ |
| 人Adx/AdR | 人CYP11A1 | 胆固醇 | 比共表达Adx/AdR的表观Vmax增加5倍以上 | [ |
| 人CYP11B1 | 11-脱氧皮质酮 | 比共表达Adx/AdR的转化率低 | [ | |
| P. putidaPdx/PdR | P. putidaP450cam | 樟脑 | 比共表达Pdx/PdR的催化速率略低 | [ |
| 比天然系统的稳态NADH氧化速率提高两倍 | [ | |||
| E. coli Fld/Fpr | B. subtilisCYP109B1 | 肉豆蔻酸 | 融合形式与非融合形式的催化活性相似 | [ |
表2 RPs融合构建实例
Table 2 Fusion construction examples of RPs
| RPs | P450酶Enzyme P450 | 底物Substrate | 融合结果Fusion results | 实例Example |
|---|---|---|---|---|
| B. megateriumP450 BM3还原酶(BMR) | 哺乳动物肝脏CYPs | 多种脂肪酸和药物 | 支持多个P450酶的催化活性,真核P450酶的溶解度明显提升 | [ |
| A. OryzaeCYP57B3 | 大豆苷元 | 支持6-OH大豆苷元最大产量为9.1 mg/L | [ | |
| JeotgalicoccusP450 OleTJE | 饱和脂肪酸 | 比Pdx/PdR的周转率高8.9倍 | [ | |
| B. subtilisCYP102A3还原酶 | B. megateriumP450 BM3血红素域 | 12-对硝基苯基十二烷酸 | 比天然酶的热稳定性明显提升 | [ |
| RhodococcusCYP116B2还原酶(RhFRED) | S. venezuelaeP450 PikC | 12元环大环内酯YC-17和14元环大环内酯那博霉素 | 比菠菜Fdx/FdR的催化活性提高约4倍 | [ |
| StreptomycesP450 TamI | 去氧糖胺糖苷修饰的碳环 | 替代菠菜Fdx/FdR实现了制备级规模的C10羟化反应 | [ | |
| A. orientalisP450Prava | 紧缩素 | 支持在产黄青霉中产生超过6 g/L的普伐他汀 | [ | |
| M. griseorubidaP450 MycG | 麦新米星 | 支持P450 MycG的常规羟化反应 | [ | |
| R. ruberCYP116B3还原酶(PFOR) | M. aquaeoleiCYP153AM.aq | 十二烷酸 | 比融合RhFRED的反应初始速率高3倍 | [ |
| T. thermophilesCYP116B46还原酶 | R. coprophilusTC-2 P450tol | 甲苯 | 比融合RhFRED的半衰期和活性分别提高20倍和2倍以上 | [ |
| 大鼠CPR | 大鼠CYP1A1 | 7-乙氧基香豆素 | 融合酶比无CPR的CYP1A1催化活性高4倍 | [ |
| ArabidopsisCPR | P. ginseng P450 PPDS | 达马烯二醇 | 比共表达CPR的催化活性高约4.5倍 | [ |
| T. cuspidataCPR | T. cuspidataCYP725A4 | 紫衫二烯 | 比共表达CPR的催化活性显著降低 | [ |
| H. tuberosusCPR | H. tuberosusCYP76B1 | 苯脲类除草剂 | 比只存在CYP76B1的除草剂耐受性降低一半以上 | [ |
| 人Adx/AdR | 人CYP11A1 | 胆固醇 | 比共表达Adx/AdR的表观Vmax增加5倍以上 | [ |
| 人CYP11B1 | 11-脱氧皮质酮 | 比共表达Adx/AdR的转化率低 | [ | |
| P. putidaPdx/PdR | P. putidaP450cam | 樟脑 | 比共表达Pdx/PdR的催化速率略低 | [ |
| 比天然系统的稳态NADH氧化速率提高两倍 | [ | |||
| E. coli Fld/Fpr | B. subtilisCYP109B1 | 肉豆蔻酸 | 融合形式与非融合形式的催化活性相似 | [ |
| [1] |
Bernhardt R. Cytochromes P450 as versatile biocatalysts[J]. J Biotechnol, 2006, 124(1): 128-145.
doi: 10.1016/j.jbiotec.2006.01.026 pmid: 16516322 |
| [2] |
Guengerich FP, Munro AW. Unusual cytochrome P450 enzymes and reactions[J]. J Biol Chem, 2013, 288(24): 17065-17073.
doi: 10.1074/jbc.R113.462275 pmid: 23632016 |
| [3] |
Zhang XW, Li SY. Expansion of chemical space for natural products by uncommon P450 reactions[J]. Nat Prod Rep, 2017, 34(9): 1061-1089.
doi: 10.1039/c7np00028f pmid: 28770915 |
| [4] |
Urlacher VB, Girhard M. Cytochrome P450 monooxygenases in biotechnology and synthetic biology[J]. Trends Biotechnol, 2019, 37(8): 882-897.
doi: S0167-7799(19)30001-0 pmid: 30739814 |
| [5] |
Li Z, Jiang YY, Guengerich FP, et al. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications[J]. J Biol Chem, 2020, 295(3): 833-849.
doi: 10.1074/jbc.REV119.008758 pmid: 31811088 |
| [6] |
Hannemann F, Bichet A, Ewen KM, et al. Cytochrome P450 systems-biological variations of electron transport chains[J]. Biochim Biophys Acta, 2007, 1770(3): 330-344.
pmid: 16978787 |
| [7] | Xu LH, Du YL. Rational and semi-rational engineering of cytochrome P450s for biotechnological applications[J]. Synth Syst Biotechnol, 2018, 3(4): 283-290. |
| [8] |
Guengerich FP. Rate-limiting steps in cytochrome P450 catalysis[J]. Biol Chem, 2002, 383(10): 1553-1564.
pmid: 12452431 |
| [9] |
Li SY, Du L, Bernhardt R. Redox partners: function modulators of bacterial P450 enzymes[J]. Trends Microbiol, 2020, 28(6): 445-454.
doi: S0966-842X(20)30048-2 pmid: 32396826 |
| [10] | Nowrouzi B, Rios-Solis L. Redox metabolism for improving whole-cell P450-catalysed terpenoid biosynthesis[J]. Crit Rev Biotechnol, 2021 Nov 8:1-25. |
| [11] |
Subedi P, Kim KH, Hong YS, et al. Enzymatic characterization and comparison of two steroid hydroxylases CYP154C3-1 and CYP154C3-2 from Streptomyces species[J]. J Microbiol Biotechnol, 2021, 31(3): 464-474.
doi: 10.4014/jmb.2010.10020 URL |
| [89] |
Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2[J]. Nat Commun, 2022, 13(1): 1265.
doi: 10.1038/s41467-022-28865-w pmid: 35273146 |
| [12] | Lin SS, Ma BB, Gao QL, et al. The 16α-hydroxylation of progesterone by cytochrome P450 107X1 from Streptomyces avermitilis[J]. Chem Biodivers, 2022, 19(5): e202200177. |
| [13] |
Jenkins CM, Waterman MR. Flavodoxin and NADPH-flavodoxin reductase from Escherichia coli support bovine cytochrome P450c17 hydroxylase activities[J]. J Biol Chem, 1994, 269(44): 27401-27408.
pmid: 7961651 |
| [14] |
Jenkins CM, Waterman MR. NADPH-flavodoxin reductase and flavodoxin from Escherichia coli: characteristics as a soluble microsomal P450 reductase[J]. Biochemistry, 1998, 37(17): 6106-6113.
pmid: 9558349 |
| [15] |
Ringle M, Khatri Y, Zapp J, et al. Application of a new versatile electron transfer system for cytochrome P450-based Escherichia coli whole-cell bioconversions[J]. Appl Microbiol Biotechnol, 2013, 97(17): 7741-7754.
doi: 10.1007/s00253-012-4612-0 URL |
| [16] |
Aliverti A, Jansen T, Zanetti G, et al. Expression in Escherichia coli of ferredoxin: NADP+ reductase from spinach. Bacterial synthesis of the holoflavoprotein and of an active enzyme form lacking the first 28 amino acid residues of the sequence[J]. Eur J Biochem, 1990, 191(3): 551-555.
pmid: 2202597 |
| [17] |
Piubelli L, Aliverti A, Bellintani F, et al. Spinach ferredoxin I: overproduction in Escherichia coli and purification[J]. Protein Expr Purif, 1995, 6(3): 298-304.
doi: 10.1006/prep.1995.1039 URL |
| [18] |
Guo C, Wu ZL. Construction and functional analysis of a whole-cell biocatalyst based on CYP108N7[J]. Enzyme Microb Technol, 2017, 106: 28-34.
doi: 10.1016/j.enzmictec.2017.06.016 URL |
| [19] |
Ewen KM, Ringle M, Bernhardt R. Adrenodoxin-a versatile ferredoxin[J]. IUBMB Life, 2012, 64(6): 506-512.
doi: 10.1002/iub.v64.6 URL |
| [20] |
Hannemann F, Virus C, Bernhardt R. Design of an Escherichia coli system for whole cell mediated steroid synthesis and molecular evolution of steroid hydroxylases[J]. J Biotechnol, 2006, 124(1): 172-181.
pmid: 16504331 |
| [21] |
Schiffer L, Anderko S, Hobler A, et al. A recombinant CYP11B1 dependent Escherichia coli biocatalyst for selective cortisol production and optimization towards a preparative scale[J]. Microb Cell Fact, 2015, 14: 25.
doi: 10.1186/s12934-015-0209-5 pmid: 25880059 |
| [22] |
Bureik M, Schiffler B, Hiraoka Y, et al. Functional expression of human mitochondrial CYP11B2 in fission yeast and identification of a new internal electron transfer protein, etp1[J]. Biochemistry, 2002, 41(7): 2311-2321.
pmid: 11841224 |
| [23] |
Ewen KM, Schiffler B, Uhlmann-Schiffler H, et al. The endogenous adrenodoxin reductase-like flavoprotein arh1 supports heterologous cytochrome P450-dependent substrate conversions in Schizosaccharomyces pombe[J]. FEMS Yeast Res, 2008, 8(3): 432-441.
doi: 10.1111/j.1567-1364.2008.00360.x URL |
| [24] |
Brixius-Anderko S, Schiffer L, Hannemann F, et al. A CYP21A2 based whole-cell system in Escherichia coli for the biotechnological production of premedrol[J]. Microb Cell Fact, 2015, 14: 135.
doi: 10.1186/s12934-015-0333-2 pmid: 26374204 |
| [25] |
Kleser M, Hannemann F, Hutter M, et al. CYP105A1 mediated 3-hydroxylation of glimepiride and glibenclamide using a recombinant Bacillus megaterium whole-cell catalyst[J]. J Biotechnol, 2012, 157(3): 405-412.
doi: 10.1016/j.jbiotec.2011.12.006 URL |
| [26] |
Zhang JJ, Lu XF, Li JJ. Conversion of fatty aldehydes into alk(a/e)nes by in vitro reconstituted cyanobacterial aldehyde-deformylating oxygenase with the cognate electron transfer system[J]. Biotechnol Biofuels, 2013, 6: 86.
doi: 10.1186/1754-6834-6-86 |
| [27] |
Ma L, Du L, Chen H, et al. Reconstitution of the in vitro activity of the cyclosporine-specific P450 hydroxylase from Sebekia benihana and development of a heterologous whole-cell biotransformation system[J]. Appl Environ Microbiol, 2015, 81(18): 6268-6275.
doi: 10.1128/AEM.01353-15 URL |
| [28] | Brill E, Hannemann F, Zapp J, et al. A new cytochrome P450 system from Bacillus megaterium DSM319 for the hydroxylation of 11-keto-β-boswellic acid(KBA)[J]. Appl Microbiol Biotechnol, 2014, 98(4): 1701-1717. |
| [29] |
Milhim M, Gerber A, Neunzig J, et al. A novel NADPH-dependent flavoprotein reductase from Bacillus megaterium acts as an efficient cytochrome P450 reductase[J]. J Biotechnol, 2016, 231: 83-94.
doi: 10.1016/j.jbiotec.2016.05.035 URL |
| [30] |
Chun YJ, Shimada T, Sanchez-Ponce R, et al. Electron transport pathway for a Streptomyces cytochrome P450: cytochrome P450 105D5-catalyzed fatty acid hydroxylation in Streptomyces coelicolor A3(2)[J]. J Biol Chem, 2007, 282(24): 17486-17500.
doi: 10.1074/jbc.M700863200 URL |
| [31] |
Ewen KM, Hannemann F, Khatri Y, et al. Genome mining in Sorangium cellulosum So Ce56: identification and characterization of the homologous electron transfer proteins of a myxobacterial cytochrome P450[J]. J Biol Chem, 2009, 284(42): 28590-28598.
doi: 10.1074/jbc.M109.021717 URL |
| [32] |
Khatri Y, Hannemann F, Ewen KM, et al. The CYPome of Sorangium cellulosum So Ce56 and identification of CYP109D1 as a new fatty acid hydroxylase[J]. Chem Biol, 2010, 17(12): 1295-1305.
doi: 10.1016/j.chembiol.2010.10.010 URL |
| [33] |
Kern F, Dier TKF, Khatri Y, et al. Highly efficient CYP167A1(EpoK)dependent epothilone B formation and production of 7-ketone epothilone D as a new epothilone derivative[J]. Sci Rep, 2015, 5: 14881.
doi: 10.1038/srep14881 |
| [34] |
Nazir KHMNH, Ichinose H, Wariishi H. Construction and application of a functional library of cytochrome P450 monooxygenases from the filamentous fungus Aspergillus oryzae[J]. Appl Environ Microbiol, 2011, 77(9): 3147-3150.
doi: 10.1128/AEM.02491-10 URL |
| [35] |
Jennewein S, Park H, DeJong JM, et al. Coexpression in yeast of Taxus cytochrome P450 reductase with cytochrome P450 oxygenases involved in Taxol biosynthesis[J]. Biotechnol Bioeng, 2005, 89(5): 588-598.
pmid: 15672381 |
| [36] |
Girhard M, Tieves F, Weber E, et al. Cytochrome P450 reductase from Candida apicola: versatile redox partner for bacterial P450s[J]. Appl Microbiol Biotechnol, 2013, 97(4): 1625-1635.
doi: 10.1007/s00253-012-4026-z pmid: 22526787 |
| [37] |
Zhang W, Liu Y, Yan J, et al. New reactions and products resulting from alternative interactions between the P450 enzyme and redox partners[J]. J Am Chem Soc, 2014, 136(9): 3640-3646.
doi: 10.1021/ja4130302 pmid: 24521145 |
| [38] |
Hiruma Y, Hass MAS, Kikui Y, et al. The structure of the cytochrome P450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography[J]. J Mol Biol, 2013, 425(22): 4353-4365.
doi: 10.1016/j.jmb.2013.07.006 pmid: 23856620 |
| [39] |
Sagadin T, Riehm JL, Milhim M, et al. Binding modes of CYP106A2 redox partners determine differences in progesterone hydroxylation product patterns[J]. Commun Biol, 2018, 1: 99.
doi: 10.1038/s42003-018-0104-9 pmid: 30271979 |
| [40] |
Khatri Y, Schifrin A, Bernhardt R. Investigating the effect of available redox protein ratios for the conversion of a steroid by a myxobacterial CYP260A1[J]. FEBS Lett, 2017, 591(8): 1126-1140.
doi: 10.1002/1873-3468.12619 pmid: 28281299 |
| [41] |
Chen CC, Min J, Zhang LL, et al. Advanced understanding of the electron transfer pathway of cytochrome P450s[J]. Chembiochem, 2021, 22(8): 1317-1328.
doi: 10.1002/cbic.v22.8 URL |
| [42] |
Zhang W, Du L, Li FW, et al. Mechanistic insights into interactions between bacterial class I P450 enzymes and redox partners[J]. ACS Catal, 2018, 8(11): 9992-10003.
doi: 10.1021/acscatal.8b02913 URL |
| [43] | Ciaramella A, Minerdi D, Gilardi G. Catalytically self-sufficient cytochromes P450 for green production of fine chemicals[J]. Rendiconti Lincei, 2017, 28(1): 169-181. |
| [44] |
Aalbers FS, Fraaije MW. Enzyme fusions in biocatalysis: coupling reactions by pairing enzymes[J]. Chembiochem, 2019, 20(1): 20-28.
doi: 10.1002/cbic.201800394 pmid: 30178909 |
| [45] |
Jung ST, Lauchli R, Arnold FH. Cytochrome P450: taming a wild type enzyme[J]. Curr Opin Biotechnol, 2011, 22(6): 809-817.
doi: 10.1016/j.copbio.2011.02.008 URL |
| [46] |
Helvig C, Capdevila JH. Biochemical characterization of rat P450 2C11 fused to rat or bacterial NADPH-P450 reductase domains[J]. Biochemistry, 2000, 39(17): 5196-5205.
pmid: 10819987 |
| [47] |
Gilardi G, Meharenna YT, Tsotsou GE, et al. Molecular Lego: design of molecular assemblies of P450 enzymes for nanobiotechnology[J]. Biosens Bioelectron, 2002, 17(1-2): 133-145.
pmid: 11742744 |
| [48] |
Fairhead M, Giannini S, Gillam EMJ, et al. Functional characterisation of an engineered multidomain human P450 2E1 by molecular Lego[J]. J Biol Inorg Chem, 2005, 10(8): 842-853.
pmid: 16283395 |
| [49] |
Dodhia VR, Fantuzzi A, Gilardi G. Engineering human cytochrome P450 enzymes into catalytically self-sufficient chimeras using molecular Lego[J]. J Biol Inorg Chem, 2006, 11(7): 903-916.
pmid: 16862439 |
| [50] |
Degregorio D, Sadeghi SJ, di Nardo G, et al. Understanding uncoupling in the multiredox centre P450 3A4-BMR model system[J]. J Biol Inorg Chem, 2011, 16(1): 109-116.
doi: 10.1007/s00775-010-0708-0 pmid: 20857167 |
| [51] |
Rua F, Sadeghi SJ, Castrignanò S, et al. Engineering Macaca fascicularis cytochrome P450 2C20 to reduce animal testing for new drugs[J]. J Inorg Biochem, 2012, 117: 277-284.
doi: 10.1016/j.jinorgbio.2012.05.017 URL |
| [52] |
Chang TS, Chao SY, Chen YC. Production of ortho-hydroxydaidzein derivatives by a recombinant strain of Pichia pastoris harboring a cytochrome P450 fusion gene[J]. Process Biochem, 2013, 48(3): 426-429.
doi: 10.1016/j.procbio.2013.02.014 URL |
| [53] |
Lu C, Shen FL, Wang SB, et al. An engineered self-sufficient biocatalyst enables scalable production of linear α-olefins from carboxylic acids[J]. ACS Catal, 2018, 8(7): 5794-5798.
doi: 10.1021/acscatal.8b01313 URL |
| [54] |
Eiben S, Bartelmäs H, Urlacher VB. Construction of a thermostable cytochrome P450 chimera derived from self-sufficient mesophilic parents[J]. Appl Microbiol Biotechnol, 2007, 75(5): 1055-1061.
pmid: 17468867 |
| [55] |
Roberts GA, Çelik A, Hunter DJB, et al. A self-sufficient cytochrome P450 with a primary structural organization that includes a flavin domain and a[2Fe-2S]redox center[J]. J Biol Chem, 2003, 278(49): 48914-48920.
doi: 10.1074/jbc.M309630200 pmid: 14514666 |
| [56] |
Li SY, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain[J]. J Am Chem Soc, 2007, 129(43): 12940-12941.
doi: 10.1021/ja075842d pmid: 17915876 |
| [57] |
Li S, Chaulagain MR, Knauff AR, et al. Selective oxidation of carbolide C-H bonds by an engineered macrolide P450 mono-oxygenase[J]. PNAS, 2009, 106(44): 18463-18468.
doi: 10.1073/pnas.0907203106 pmid: 19833867 |
| [58] |
Carlson JC, Li SY, Gunatilleke SS, et al. Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes[J]. Nat Chem, 2011, 3(8): 628-633.
doi: 10.1038/nchem.1087 pmid: 21778983 |
| [59] |
McLean KJ, Hans M, Meijrink B, et al. Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum[J]. Proc Natl Acad Sci USA, 2015, 112(9): 2847-2852.
doi: 10.1073/pnas.1419028112 URL |
| [60] |
Sabbadin F, Hyde R, Robin A, et al. LICRED: a versatile drop-in vector for rapid generation of redox-self-sufficient cytochrome P450s[J]. Chembiochem, 2010, 11(7): 987-994.
doi: 10.1002/cbic.201000104 pmid: 20425752 |
| [61] |
Hoffmann SM, Weissenborn MJ, Gricman Ł, et al. The impact of linker length on P450 fusion constructs: activity, stability and coupling[J]. ChemCatChem, 2016, 8(8): 1591-1597.
doi: 10.1002/cctc.201501397 URL |
| [62] |
Chen CC, Dai M, Zhang LL, et al. Molecular basis for a toluene monooxygenase to govern substrate selectivity[J]. ACS Catal, 2022, 12(5): 2831-2839.
doi: 10.1021/acscatal.1c05845 URL |
| [63] |
Murakami H, Yabusaki Y, Sakaki T, et al. A genetically engineered P450 monooxygenase: construction of the functional fused enzyme between rat cytochrome P450c and NADPH-cytochrome P450 reductase[J]. DNA, 1987, 6(3): 189-197.
doi: 10.1089/dna.1987.6.189 URL |
| [64] |
Shet MS, Fisher CW, Arlotto MP, et al. Purification and enzymatic properties of a recombinant fusion protein expressed in Escherichia coli containing the domains of bovine P450 17A and rat NADPH-P450 reductase[J]. Arch Biochem Biophys, 1994, 311(2): 402-417.
pmid: 8203904 |
| [65] |
Zhao FL, Bai P, Liu T, et al. Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae[J]. Biotechnol Bioeng, 2016, 113(8): 1787-1795.
doi: 10.1002/bit.v113.8 URL |
| [66] |
Biggs BW, Lim CG, Sagliani K, et al. Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli[J]. Proc Natl Acad Sci USA, 2016, 113(12): 3209-3214.
doi: 10.1073/pnas.1515826113 URL |
| [67] |
Didierjean L, Gondet L, Perkins R, et al. Engineering herbicide metabolism in tobacco and Arabidopsis with CYP76B1, a cytochrome P450 enzyme from Jerusalem artichoke[J]. Plant Physiol, 2002, 130(1): 179-189.
pmid: 12226498 |
| [68] |
Harikrishna JA, Black SM, Szklarz GD, et al. Construction and function of fusion enzymes of the human cytochrome P450scc system[J]. DNA Cell Biol, 1993, 12(5): 371-379.
pmid: 8517924 |
| [69] |
Cao PR, Bülow H, Dumas B, et al. Construction and characterization of a catalytic fusion protein system: P-45011β-adrenodoxin reductase-adrenodoxin[J]. Biochim Biophys Acta Protein Struct Mol Enzymol, 2000, 1476(2): 253-264.
doi: 10.1016/S0167-4838(99)00243-5 URL |
| [70] |
Sibbesen O, de Voss JJ, de Montellano PRO. Putidaredoxin reductase-putidaredoxin-cytochrome P450cam triple fusion protein[J]. J Biol Chem, 1996, 271(37): 22462-22469.
doi: 10.1074/jbc.271.37.22462 pmid: 8798411 |
| [71] |
Johnson EO, Wong LL. Partial fusion of a cytochrome P450 system by carboxy-terminal attachment of putidaredoxin reductase to P450cam(CYP101A1)[J]. Catal Sci Technol, 2016, 6(20): 7549-7560.
doi: 10.1039/C6CY01042C pmid: 28944003 |
| [72] |
Strushkevich N, MacKenzie F, Cherkesova T, et al. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system[J]. Proc Natl Acad Sci USA, 2011, 108(25): 10139-10143.
doi: 10.1073/pnas.1019441108 pmid: 21636783 |
| [73] |
Bakkes PJ, Biemann S, Bokel A, et al. Design and improvement of artificial redox modules by molecular fusion of flavodoxin and flavodoxin reductase from Escherichia coli[J]. Sci Rep, 2015, 5: 12158.
doi: 10.1038/srep12158 |
| [74] | Robin A, Roberts GA, Kisch J, et al. Engineering and improvement of the efficiency of a chimeric[P450cam-RhFRed reductase domain]enzyme[J]. Chem Commun, 2009(18): 2478. |
| [75] |
Belsare KD, Ruff AJ, Martinez R, et al. P-LinK: a method for generating multicomponent cytochrome P450 fusions with variable linker length[J]. BioTechniques, 2014, 57(1): 13-20.
doi: 10.2144/000114187 pmid: 25005689 |
| [76] |
Hlavica P, Schulze J, Lewis DFV. Functional interaction of cytochrome P450 with its redox partners: a critical assessment and update of the topology of predicted contact regions[J]. J Inorg Biochem, 2003, 96(2-3): 279-297.
pmid: 12888264 |
| [77] |
Tripathi S, Li HY, Poulos TL. Structural basis for effector control and redox partner recognition in cytochrome P450[J]. Science, 2013, 340(6137): 1227-1230.
doi: 10.1126/science.1235797 pmid: 23744947 |
| [78] |
Sevrioukova IF, Poulos TL, Churbanova IY. Crystal structure of the putidaredoxin Reductase·Putidaredoxin electron transfer complex[J]. J Biol Chem, 2010, 285(18): 13616-13620.
doi: 10.1074/jbc.M110.104968 pmid: 20179327 |
| [79] |
Müller A, Müller JJ, Muller YA, et al. New aspects of electron transfer revealed by the crystal structure of a truncated bovine adrenodoxin, Adx(4-108)[J]. Structure, 1998, 6(3): 269-280.
pmid: 9551550 |
| [80] |
Uhlmann H, Kraft R, Bernhardt R. C-terminal region of adrenodoxin affects its structural integrity and determines differences in its electron transfer function to cytochrome P-450[J]. J Biol Chem, 1994, 269(36): 22557-22564.
pmid: 8077204 |
| [81] | Bell SG, McMillan JHC, Yorke JA, et al. Tailoring an alien ferredoxin to support native-like P450 monooxygenase activity[J]. Chem Commun(Camb), 2012, 48(95): 11692-11694. |
| [82] |
Sagadin T, Riehm J, Putkaradze N, et al. Novel approach to improve progesterone hydroxylation selectivity by CYP106A2 via rational design of adrenodoxin binding[J]. FEBS J, 2019, 286(6): 1240-1249.
doi: 10.1111/febs.14722 pmid: 30537187 |
| [83] |
Kabumoto H, Miyazaki K, Arisawa A. Directed evolution of the actinomycete cytochrome P450 MoxA(CYP105)for enhanced activity[J]. Biosci Biotechnol Biochem, 2009, 73(9): 1922-1927.
doi: 10.1271/bbb.90013 URL |
| [84] |
Ba LN, Li P, Zhang H, et al. Semi-rational engineering of cytochrome P450sca-2 in a hybrid system for enhanced catalytic activity: insights into the important role of electron transfer[J]. Biotechnol Bioeng, 2013, 110(11): 2815-2825.
doi: 10.1002/bit.24960 pmid: 23737252 |
| [85] |
Poulos TL, Follmer AH. Updating the paradigm: redox partner binding and conformational dynamics in cytochromes P450[J]. Acc Chem Res, 2022, 55(3): 373-380.
doi: 10.1021/acs.accounts.1c00632 URL |
| [86] |
Hlavica P. Assembly of non-natural electron transfer conduits in the cytochrome P450 system: a critical assessment and update of artificial redox constructs amenable to exploitation in biotechnological areas[J]. Biotechnol Adv, 2009, 27(2): 103-121.
doi: 10.1016/j.biotechadv.2008.10.001 pmid: 18976700 |
| [87] |
Mellor SB, Vavitsas K, Nielsen AZ, et al. Photosynthetic fuel for heterologous enzymes: the role of electron carrier proteins[J]. Photosynth Res, 2017, 134(3): 329-342.
doi: 10.1007/s11120-017-0364-0 URL |
| [88] |
Hirakawa H, Kamiya N, Tanaka T, et al. Intramolecular electron transfer in a cytochrome P450cam system with a site-specific branched structure[J]. Protein Eng Des Sel, 2007, 20(9): 453-459.
pmid: 17827502 |
| [1] | 郁慧丽, 李爱涛. 细胞色素P450酶在香精香料绿色生物合成中的应用[J]. 生物技术通报, 2023, 39(4): 24-37. |
| [2] | 魏倩, 刘小宁, 赵洁. 2-十三烷酮胁迫下棉铃虫FoxAl调控CYP6B6的表达[J]. 生物技术通报, 2022, 38(5): 84-92. |
| [3] | 袁林, 黄朝, 曾静, 郭建军, 张婷, 吕珺,. 植酸酶YiAPPA与生淀粉结合域SBD融合酶的构建及酶学性质分析[J]. 生物技术通报, 2018, 34(3): 200-207. |
| [4] | P.N.F.Thoneley. 对固氮酶的新见解[J]. , 1994, 0(01): 8-9. |
| [5] | 李思经;. 一种测定植物细胞存活力的新方法[J]. , 1988, 0(11): 9-9. |
| [6] | . 农药[J]. , 1985, 0(02): 67-71. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||