








生物技术通报 ›› 2023, Vol. 39 ›› Issue (7): 288-297.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1430
周振超1( ), 郑吉2, 帅馨怡1, 林泽俊1, 陈红1(
), 郑吉2, 帅馨怡1, 林泽俊1, 陈红1( )
)
收稿日期:2022-11-21
出版日期:2023-07-26
发布日期:2023-08-17
通讯作者:
陈红,女,博士,教授,研究方向:环境抗生素耐药;E-mail: chen_hong@zju.edu.cn作者简介:周振超,男,博士,助理研究员,研究方向:环境抗生素耐药;E-mail: zhouzc@zju.edu.cn
基金资助:
ZHOU Zhen-chao1( ), ZHENG Ji2, SHUAI Xin-yi1, LIN Ze-jun1, CHEN Hong1(
), ZHENG Ji2, SHUAI Xin-yi1, LIN Ze-jun1, CHEN Hong1( )
)
Received:2022-11-21
Published:2023-07-26
Online:2023-08-17
摘要:
抗生素抗性基因(antibiotic resistance genes, ARGs)是一种新污染物,其在环境中的传播加剧了抗生素耐药问题。但是,关于人体和水环境中共享ARGs的分布及潜在宿主知之甚少。为此,本文使用高通量荧光定量PCR和16S rRNA基因测序,分析来自中国城郊地区的人体和水环境中的ARGs结构和细菌群落特征。在粪便、皮肤和水样中共确定了人体和水环境之间的70个共享ARGs、7个共享移动遗传元件(mobile genetic elements, MGEs)和58个共享细菌。粪便中占主导地位的共享ARGs为四环素和MLSB抗性基因。此外,通过LEfSe(linear discriminant analysis effect size)确定了20种细菌生物标志物。通过网络分析揭示了共享ARGs、MGEs与共享细菌具有显著相关(P < 0.05),表明这些细菌可能是ARGs的潜在宿主并且在人体和环境之间转移。本研究揭示了饮用水、粪便、皮肤、污水和地表水样本中共享的ARGs。研究结果更好地理解了共享ARGs和细菌在环境中的共同发生和传播,揭示了城郊地区人体与环境中ARGs的潜在宿主。
周振超, 郑吉, 帅馨怡, 林泽俊, 陈红. 高通量分析人类粪便、皮肤和水环境中共享抗生素抗性基因的分布[J]. 生物技术通报, 2023, 39(7): 288-297.
ZHOU Zhen-chao, ZHENG Ji, SHUAI Xin-yi, LIN Ze-jun, CHEN Hong. High-throughput Profiling and Analysis of Shared Antibiotic Resistance Genes in Human Feces, Skin and Water Environments[J]. Biotechnology Bulletin, 2023, 39(7): 288-297.
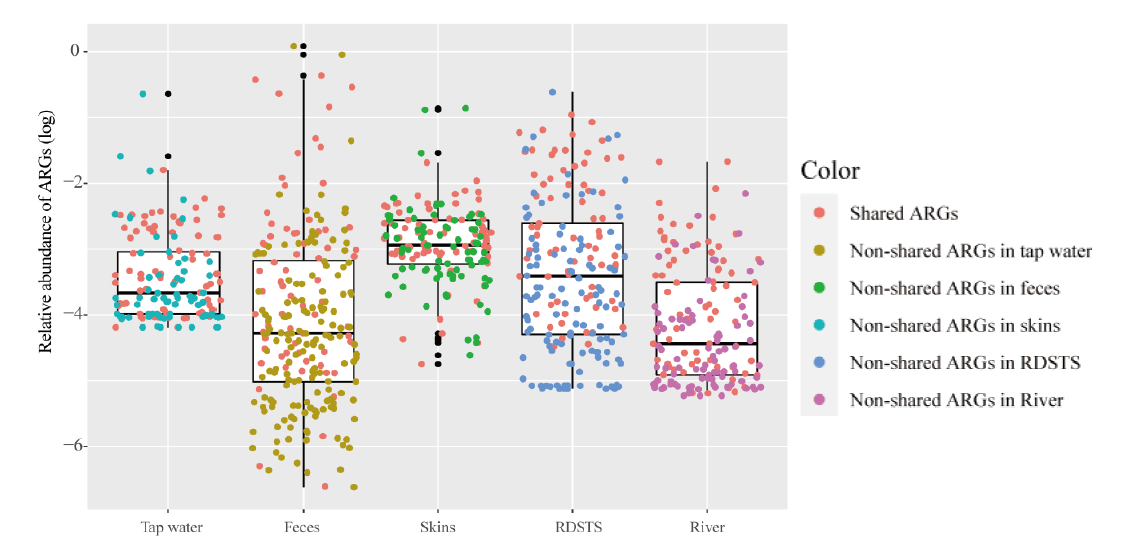
图1 人体和水环境样品中抗生素抗性基因相对丰度 红色圆点代表了70个共享抗生素抗性基因。误差棒代表了最大最小值,中线代表了中位值
Fig. 1 Relative abundances of ARGs in humans and water samples Red points indicate commonly shared ARGs in human feces, skin and water(n =70). Error bars indicate min/max values, and centre bars indicate median

图4 基于LEfSe区分人类粪便、皮肤和水环境中差异特征微生物分类群 A:在系统发育树中显示了门到科类别的分类群(Kruskal-Wallis 和成对 Wilcoxon 秩和检验的 alpha 值 < 0.05,LDA 效应大小≥ 4.0),圆圈大小与分类群丰度成正比。B,C:人类粪便、皮肤和水微生物群中区分微生物门(B)和科(C)的相对丰度。仅显示了一个类别中平均相对丰度≥ 1% 的分类群。误差条代表标准偏差,中线代表中位值,黑点代表异常值。
Fig. 4 Taxa discriminates among human feces, skin and water microbiota as determined by LEfSe A: Kingdom- to family-level taxa were shown in the phylogenetic tree. Colored taxa are discriminative between categories(alpha value of < 0.05 for both the Kruskal-Wallis and pairwise Wilcoxon rank-sum tests, and LDA effect size of ≥ 4.0), circle size is proportional to the taxa abundance. B, C: Relative abundances of discriminative microbial phyla(B)and families(C)in human feces, skin and water microbiota. Only taxa with a mean relative abundance of ≥ 1% in one category are shown. Error bars indicate standard deviation(s. d.), centre bars indicate median, and black points indicate outliers
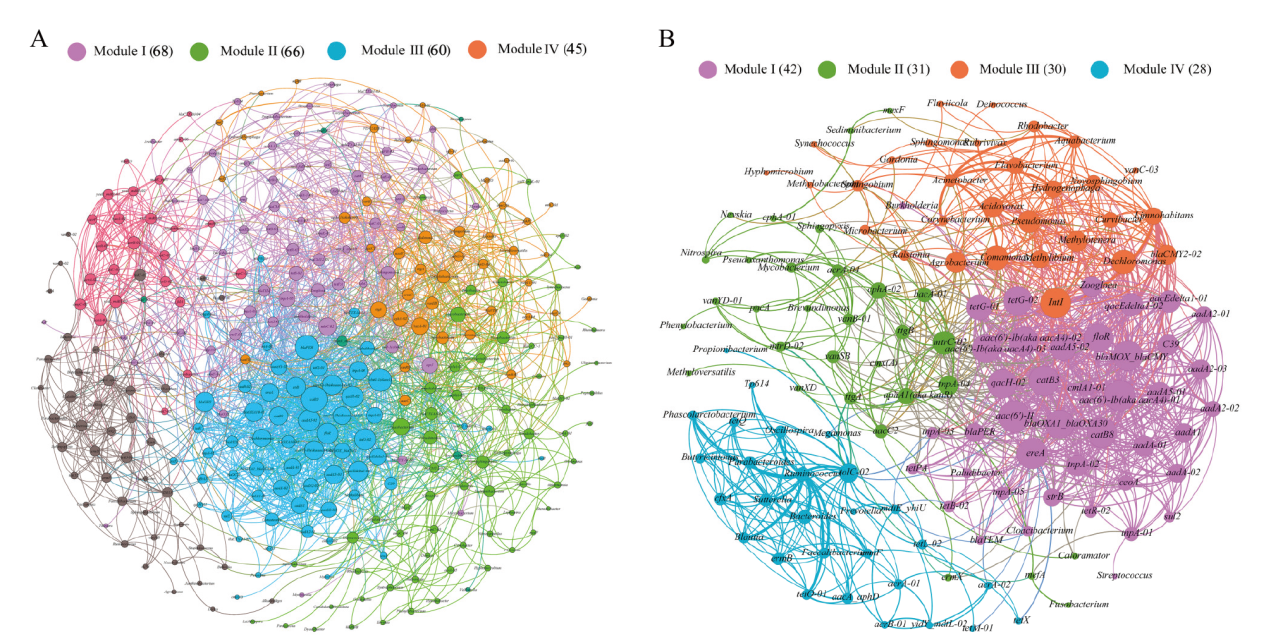
图6 所有检测到的 ARGs、MGEs和细菌的网络分析(A)和共享 ARGs、MGEs和细菌的网络分析(B) 节点根据模块化等级着色。连接表示强(Spearman 相关系数(ρ)>0.7)和显著(P 值<0.01)相关性。根据连接数(即度)加权的节点大小和根据相关系数加权的边
Fig. 6 Network analysis of all detected ARGs, MGEs and bacteria(A)and shared ARGs, MGEs and bacteria(B) The nodes were colored according to modularity class. A connection represents a strong(Spearman's correlation coefficient(ρ)>0.7)and significant(P-value<0.01)correlation. Node size weighted according to the number of connections(that is the degree)and edges weighted according to the correlation coefficient
| [1] |
Pruden A, Pei RT, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado[J]. Environ Sci Technol, 2006, 40(23): 7445-7450.
pmid: 17181002 |
| [2] |
Shi P, Jia SY, Zhang XX, et al. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water[J]. Water Res, 2013, 47(1): 111-120.
doi: 10.1016/j.watres.2012.09.046 pmid: 23084468 |
| [3] |
Xu LK, Ouyang WY, Qian YY, et al. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems[J]. Environ Pollut, 2016, 213: 119-126.
doi: S0269-7491(16)30116-6 pmid: 26890482 |
| [4] |
Bai XH, Ma XL, Xu FM, et al. The drinking water treatment process as a potential source of affecting the bacterial antibiotic resistance[J]. Sci Total Environ, 2015, 533: 24-31.
doi: 10.1016/j.scitotenv.2015.06.082 URL |
| [5] |
Chen H, Zhang MM. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in Eastern China[J]. Environ Int, 2013, 55: 9-14.
doi: 10.1016/j.envint.2013.01.019 pmid: 23454279 |
| [6] |
Mao DQ, Yu S, Rysz M, et al. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants[J]. Water Res, 2015, 85: 458-466.
doi: 10.1016/j.watres.2015.09.010 pmid: 26372743 |
| [7] |
Di Cesare A, Eckert EM, D’Urso S, et al. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants[J]. Water Res, 2016, 94: 208-214.
doi: S0043-1354(16)30108-7 pmid: 26945964 |
| [8] |
Zheng J, Gao RX, Wei YY, et al. High-throughput profiling and analysis of antibiotic resistance genes in East Tiaoxi River, China[J]. Environ Pollut, 2017, 230: 648-654.
doi: S0269-7491(17)31437-9 pmid: 28715769 |
| [9] |
Xu J, Xu Y, Wang HM, et al. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river[J]. Chemosphere, 2015, 119: 1379-1385.
doi: S0045-6535(14)00244-6 pmid: 24630248 |
| [10] |
Luo Y, Mao DQ, Rysz M, et al. Trends in antibiotic resistance genes occurrence in the Haihe River, China[J]. Environ Sci Technol, 2010, 44(19): 7220-7225.
doi: 10.1021/es100233w pmid: 20509603 |
| [11] |
Pal C, Bengtsson-Palme J, Kristiansson E, et al. The structure and diversity of human, animal and environmental resistomes[J]. Microbiome, 2016, 4(1): 54.
pmid: 27717408 |
| [12] |
Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography[J]. Nature, 2012, 486(7402): 222-227.
doi: 10.1038/nature11053 |
| [13] | Pärnänen K, Karkman A, Hultman J, et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements[J]. Nat Commun, 2018, 9, 3891. |
| [14] |
Schommer NN, Gallo RL. Structure and function of the human skin microbiome[J]. Trends Microbiol, 2013, 21(12): 660-668.
doi: 10.1016/j.tim.2013.10.001 pmid: 24238601 |
| [15] |
Yang Y, Li B, Zou SC, et al. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach[J]. Water Res, 2014, 62: 97-106.
doi: 10.1016/j.watres.2014.05.019 pmid: 24937359 |
| [16] |
Zheng J, Zhou ZC, Wei YY, et al. High-throughput profiling of seasonal variations of antibiotic resistance gene transport in a peri-urban river[J]. Environ Int, 2018, 114: 87-94.
doi: S0160-4120(17)31942-6 pmid: 29499451 |
| [17] |
Jia SY, Shi P, Hu Q, et al. Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination[J]. Environ Sci Technol, 2015, 49(20): 12271-12279.
doi: 10.1021/acs.est.5b03521 pmid: 26397118 |
| [18] |
Rizzo L, Manaia C, Merlin C, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review[J]. Sci Total Environ, 2013, 447: 345-360.
doi: 10.1016/j.scitotenv.2013.01.032 URL |
| [19] |
Guo JH, Li J, Chen H, et al. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements[J]. Water Res, 2017, 123: 468-478.
doi: S0043-1354(17)30565-1 pmid: 28689130 |
| [20] |
Su JQ, An XL, Li B, et al. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China[J]. Microbiome, 2017, 5(1): 84.
doi: 10.1186/s40168-017-0298-y URL |
| [21] |
Peng F, Isabwe A, Guo YY, et al. An extensively shared antibiotic resistome among four seasons suggests management prioritization in a subtropical riverine ecosystem[J]. Sci Total Environ, 2019, 673: 533-540.
doi: 10.1016/j.scitotenv.2019.04.031 URL |
| [22] | Zhou ZC, Lin ZJ, Shuai XY, et al. Temporal variation and sharing of antibiotic resistance genes between water and wild fish gut in a peri-urban river[J]. J Environ Sci(China), 2021, 103: 12-19. |
| [23] |
Redondo-Salvo S, Fernández-López R, Ruiz R, et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids[J]. Nat Commun, 2020, 11(1): 3602.
doi: 10.1038/s41467-020-17278-2 pmid: 32681114 |
| [24] |
Ma LP, Xia Y, Li B, et al. Metagenomic assembly reveals hosts of antibiotic resistance genes and the shared resistome in pig, chicken, and human feces[J]. Environ Sci Technol, 2016, 50(1): 420-427.
doi: 10.1021/acs.est.5b03522 pmid: 26650334 |
| [25] |
Zhu YG, Gillings M, Simonet P, et al. Human dissemination of genes and microorganisms in Earth's Critical Zone[J]. Glob Change Biol, 2018, 24(4): 1488-1499.
doi: 10.1111/gcb.2018.24.issue-4 URL |
| [26] |
Zhou ZC, Feng WQ, Han Y, et al. Prevalence and transmission of antibiotic resistance and microbiota between humans and water environments[J]. Environ Int, 2018, 121(Pt 2): 1155-1161.
doi: 10.1016/j.envint.2018.10.032 URL |
| [27] |
Oh J, Byrd AL, Park M, et al. Temporal stability of the human skin microbiome[J]. Cell, 2016, 165(4): 854-866.
doi: 10.1016/j.cell.2016.04.008 pmid: 27153496 |
| [28] |
Oh J, Byrd AL, Deming C, et al. Biogeography and individuality shape function in the human skin metagenome[J]. Nature, 2014, 514(7520): 59-64.
doi: 10.1038/nature13786 |
| [29] |
Shuai XY, Sun YJ, Meng LX, et al. Dissemination of antibiotic resistance genes in swimming pools and implication for human skin[J]. Sci Total Environ, 2021, 794: 148693.
doi: 10.1016/j.scitotenv.2021.148693 URL |
| [30] |
Turner S, Pryer KM, Miao VP, et al. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis[J]. J Eukaryot Microbiol, 1999, 46(4): 327-338.
doi: 10.1111/j.1550-7408.1999.tb04612.x pmid: 10461381 |
| [31] |
Cui EP, Wu Y, Zuo YR, et al. Effect of different biochars on antibiotic resistance genes and bacterial community during chicken manure composting[J]. Bioresour Technol, 2016, 203: 11-17.
doi: 10.1016/j.biortech.2015.12.030 URL |
| [32] |
Wu Y, Cui EP, Zuo YR, et al. Influence of two-phase anaerobic digestion on fate of selected antibiotic resistance genes and class I integrons in municipal wastewater sludge[J]. Bioresour Technol, 2016, 211: 414-421.
doi: 10.1016/j.biortech.2016.03.086 URL |
| [33] | Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation[J]. Genome Biol, 2011, 12(6): R60. |
| [34] | Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks[J]. Proc Int AAAI Conf Web Soc Media, 2009, 3(1): 361-362. |
| [35] |
Zhu YG, Zhao Y, Li B, et al. Continental-scale pollution of estuaries with antibiotic resistance genes[J]. Nat Microbiol, 2017, 2: 16270.
doi: 10.1038/nmicrobiol.2016.270 |
| [36] |
Rodriguez-Mozaz S, Chamorro S, Marti E, et al. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river[J]. Water Res, 2015, 69: 234-242.
doi: S0043-1354(14)00791-X pmid: 25482914 |
| [37] |
Pehrsson EC, Tsukayama P, Patel S, et al. Interconnected microbiomes and resistomes in low-income human habitats[J]. Nature, 2016, 533(7602): 212-216.
doi: 10.1038/nature17672 |
| [38] |
You XX, Wu D, Wei HW, et al. Fluoroquinolones and β-lactam antibiotics and antibiotic resistance genes in autumn leachates of seven major municipal solid waste landfills in China[J]. Environ Int, 2018, 113: 162-169.
doi: S0160-4120(17)31954-2 pmid: 29425900 |
| [39] |
Zhang QQ, Ying GG, Pan CG, et al. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance[J]. Environ Sci Technol, 2015, 49(11): 6772-6782.
doi: 10.1021/acs.est.5b00729 URL |
| [40] |
Karkman A, Do TT, Walsh F, et al. Antibiotic-resistance genes in waste water[J]. Trends Microbiol, 2018, 26(3): 220-228.
doi: S0966-842X(17)30210-X pmid: 29033338 |
| [41] |
Huang XP, Mo CH, Yu J, et al. Variations in microbial community and ciprofloxacin removal in rhizospheric soils between two cultivars of Brassica parachinensis L[J]. Sci Total Environ, 2017, 603/604: 66-76.
doi: 10.1016/j.scitotenv.2017.06.040 URL |
| [42] |
Jiao YN, Zhou ZC, Chen T, et al. Biomarkers of antibiotic resistance genes during seasonal changes in wastewater treatment systems[J]. Environ Pollut, 2018, 234: 79-87.
doi: 10.1016/j.envpol.2017.11.048 URL |
| [43] |
Zhou ZC, Zheng J, Wei YY, et al. Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters[J]. Environ Sci Pollut Res, 2017, 24(30): 23753-23762.
doi: 10.1007/s11356-017-0032-0 |
| [44] |
Zhang SY, Su JQ, Sun GX, et al. Land scale biogeography of arsenic biotransformation genes in estuarine wetland[J]. Environ Microbiol, 2017, 19(6): 2468-2482.
doi: 10.1111/1462-2920.13775 URL |
| [45] | WHO. World Health Organization. https://www.who.int/zh/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed[M]. 2017. |
| [46] |
Li B, Yang Y, Ma LP, et al. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes[J]. ISME J, 2015, 9(11): 2490-2502.
doi: 10.1038/ismej.2015.59 pmid: 25918831 |
| [1] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [2] | 孙海航, 官会林, 王旭, 王童, 李泓霖, 彭文洁, 刘柏桢, 樊芳玲. 生物炭对三七连作土壤性质及真菌群落的影响[J]. 生物技术通报, 2023, 39(2): 221-231. |
| [3] | 刘金升, 陈振娅, 霍毅欣, 郭淑元. FACS技术在酶定向进化中的应用[J]. 生物技术通报, 2023, 39(10): 93-106. |
| [4] | 胡雪莹, 张越, 郭雅杰, 仇天雷, 高敏, 孙兴滨, 王旭明. 不同施肥处理农田土壤中噬菌体与细菌携带抗生素抗性基因的比较[J]. 生物技术通报, 2022, 38(9): 116-126. |
| [5] | 王子夜, 王志刚, 阎爱华. 不同树龄桑根际土壤原生生物群落组成多样性[J]. 生物技术通报, 2022, 38(8): 206-215. |
| [6] | 陈天赐, 武少兰, 杨国辉, 江丹霞, 江玉姬, 陈炳智. 无柄灵芝醇提物对小鼠睡眠及肠道菌群的影响[J]. 生物技术通报, 2022, 38(8): 225-232. |
| [7] | 钟辉, 刘亚军, 王滨花, 和梦洁, 吴兰. 分析方法对细菌群落16S rRNA基因扩增测序分析结果的影响[J]. 生物技术通报, 2022, 38(6): 81-92. |
| [8] | 朱秋雨, 段绪果. L-天冬氨酸-α-脱羧酶的重组表达、定点突变及高通量检测方法的建立[J]. 生物技术通报, 2022, 38(5): 269-278. |
| [9] | 杨青青, 唐家琪, 张昌泉, 高继平, 刘巧泉. KASP标记技术在主要农作物中的应用及展望[J]. 生物技术通报, 2022, 38(4): 58-71. |
| [10] | 赵林艳, 官会林, 向萍, 李泽诚, 柏雨龙, 宋洪川, 孙世中, 徐武美. 白及根腐病植株根际土壤微生物群落组成特征分析[J]. 生物技术通报, 2022, 38(2): 67-74. |
| [11] | 陈宇捷, 郑华宝, 周昕彦. 改良高通量测序技术揭示除藻剂对藻类群落的影响[J]. 生物技术通报, 2022, 38(11): 70-79. |
| [12] | 曹修凯, 王珊, 葛玲, 张卫博, 孙伟. 染色体外环形DNA研究进展及其在畜禽育种中的应用[J]. 生物技术通报, 2022, 38(1): 247-257. |
| [13] | 张雪, 谭玉萌, 蒋海霞, 杨广宇. 基于单细胞超高通量筛选的α-1,2-岩藻糖基转移酶定向进化[J]. 生物技术通报, 2022, 38(1): 289-298. |
| [14] | 毛婷, 牛永艳, 郑群, 杨涛, 穆永松, 祝英, 季彬, 王治业. 菌剂对苜蓿青贮发酵品质及微生物群落的影响[J]. 生物技术通报, 2021, 37(9): 86-94. |
| [15] | 唐蝶, 周倩. 植物基因组组装技术研究进展[J]. 生物技术通报, 2021, 37(6): 1-12. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||