








生物技术通报 ›› 2023, Vol. 39 ›› Issue (8): 241-250.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1562
李博1,3( ), 刘合霞1, 陈宇玲1, 周兴文2, 朱宇林1,3(
), 刘合霞1, 陈宇玲1, 周兴文2, 朱宇林1,3( )
)
收稿日期:2022-12-29
出版日期:2023-08-26
发布日期:2023-09-05
通讯作者:
朱宇林,男,博士,教授,研究方向:植物资源利用及种质创新;E-mail: gxzyl@163.com作者简介:李博,男,博士,副教授,研究方向:金花茶的资源利用;E-mail: lbshaojianbo@ylu.edu.cn
基金资助:
LI Bo1,3( ), LIU He-xia1, CHEN Yu-ling1, ZHOU Xing-wen2, ZHU Yu-lin1,3(
), LIU He-xia1, CHEN Yu-ling1, ZHOU Xing-wen2, ZHU Yu-lin1,3( )
)
Received:2022-12-29
Published:2023-08-26
Online:2023-09-05
摘要:
为探究CnbHLH79转录因子在金花茶花色形成中的作用机理,克隆了金花茶CnbHLH79转录因子的编码序列,对其进行生物信息学和亚细胞定位分析,利用荧光定量PCR技术分析了CnbHLH79转录因子在金花茶不同组织,以及不同发育阶段的花瓣中的表达模式,并分析CnbHLH79转录因子的表达量与色相b*、类黄酮物质含量的相关关系。研究结果表明,CnbHLH79转录因子的开放阅读框长度为855 bp,共编码284个氨基酸,具有bHLH_AtBPE_like保守结构域。系统进化分析发现,金花茶CnbHLH79蛋白与茶树bHLH79蛋白的亲缘关系最近。亚细胞定位分析显示,CnbHLH79转录因子主要在细胞核中发挥功能。对CnbHLH79转录因子进行实时荧光定量PCR分析发现,该转录因子在金花茶的根、花等组织中表达量较高;还发现在花朵开放过程中,CnbHLH79转录因子的表达量总体呈先高后低,逐步下降的趋势;此外,CnbHLH79的表达量与色相b*值、槲皮素-7-O-葡糖苷的相关性系数分别为-0.92、-0.7,它们之间的负相关性较高。本研究将为阐明CnbHLH79转录因子在金花茶中调控类黄酮的合成机制,以及调控花瓣显黄色的作用机理奠定基础。
李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250.
LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima[J]. Biotechnology Bulletin, 2023, 39(8): 241-250.

图1 用于荧光定量表达的金花茶不同组织 从左到右分别是侧根、茎、叶;不同的开花时期分别为幼蕾期(A)、初蕾期(B)、显色期(C)、半开期(D)、盛开期(E)
Fig. 1 Different cultures of C. nitidissima for fluorescence quantitative analysis The cultures of C. nitidissima are lateral root, stem and leave from left to right. The different flowering stages are young bud(A), early bud(B), chromogenic stages of flowering(C), the half-opening stage(D)and blooming stage(E)
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Use |
|---|---|---|
| CnbHLH79-F | GATCCTCCGATAATCAATTTGACCTCTTTC | 克隆Clone |
| CnbHLH79-R | TGCTGCTCTATCAAAACTGCTACCAAC | |
| CnbHLH79-F | GGAACACCAACATTTGATGC | 定量表达Quantitative expression |
| CnbHLH79-R | ATGAAGCCATTCGGTTTGT | |
| CnbHLH79-F | ACTAGGGTCTCGCACCATGGATCCTCCGATAATCAATTTGACCTCTTTC | 亚细胞定位Subcellular localization |
| CnbHLH79-R | ACTAGGGTCTCTCGCC TGCTGCTCTATCAAAACTGCTACCAAC | |
| 18S rRNA-F | CAACCATAAACGATGCCGA | 内参基因Reference gene |
| 18S rRNA-R | AGCCTTGCGACCATACTCC |
表1 CnbHLH79转录因子克隆、亚细胞定位以及表达定量所用引物
Table 1 Primers used for cloning, subcellular localization and quantitative analysis of CnbHLH79
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Use |
|---|---|---|
| CnbHLH79-F | GATCCTCCGATAATCAATTTGACCTCTTTC | 克隆Clone |
| CnbHLH79-R | TGCTGCTCTATCAAAACTGCTACCAAC | |
| CnbHLH79-F | GGAACACCAACATTTGATGC | 定量表达Quantitative expression |
| CnbHLH79-R | ATGAAGCCATTCGGTTTGT | |
| CnbHLH79-F | ACTAGGGTCTCGCACCATGGATCCTCCGATAATCAATTTGACCTCTTTC | 亚细胞定位Subcellular localization |
| CnbHLH79-R | ACTAGGGTCTCTCGCC TGCTGCTCTATCAAAACTGCTACCAAC | |
| 18S rRNA-F | CAACCATAAACGATGCCGA | 内参基因Reference gene |
| 18S rRNA-R | AGCCTTGCGACCATACTCC |

图3 金花茶CnbHLH79蛋白功能结构域预测及氨基酸多重序列比对 A:蛋白功能结构域预测(bHLH_AtBPE_like:保守结构域;bHLH_SF:超基因家族);B:氨基酸序列同源性比较;红色划线区域:bHLH_SF基因家族保守结构域,bHLH79蛋白在该区域的氨基酸序列高度保守;α1和α2:α-螺旋元件;CnbHLH79:金花茶bHLH79转录因子;AtbHLH79:拟南芥bHLH79转录因子(AT5G62610.1);CsbHLH79_like:茶树bHLH79转录因子(XP_028119408.1);ArBPEp:褐枝猕猴桃BPEp转录因子(GFS32687.1)
Fig. 3 Prediting functional domain of protein CnbHLH79 and amino acid multiple sequence aligning in C. nitidissima A: Protein functional domains prediction(bHLH_AtBPE_like: conserved domains; bHLH_SF: superfamily). B: Homology comparison of putative amino acids sequence. The sequence underlined in red: conserved domains of bHLH_SF superfamily; conserved amino acids of bHLH_SF superfamily contained by bHLH79 protein; α1 and α2: α-helix; CnbHLH79: bHLH79 transcription factor of C. nitidissima; AtbHLH79: bHLH79 transcription factor of Arabidopsis thaliana(AT5G62610.1); CsbHLH79_like: bHLH79 transcription factor of C. sinensis(XP_028119408.1); ArBPEp: BPEp transcription factor of A. rufa(GFS32687.1)
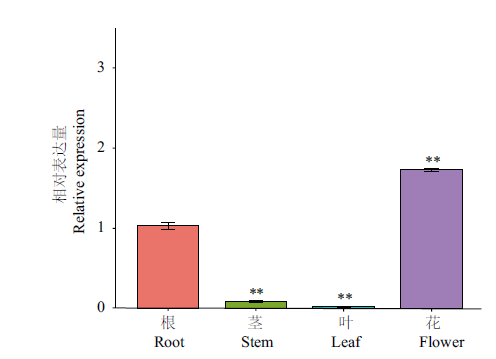
图7 CnbHLH79在不同组织中的基因表达量 **代表茎、叶、花的表达量相对于根,差异极显著(P<0.01);内参基因:18S rRNA;n=3
Fig. 7 Relative expressions of CnbHLH79 in different culture ** indicates extremely significant difference in the expression of stem, leave and flower compared with root(P<0.01). Reference gene: 18S rRNA; n=3
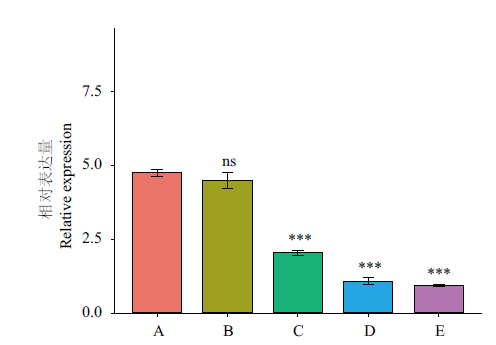
图8 CnbHLH79转录因子在不同开花时期的表达量 A:幼蕾期;B:初蕾期;C:显色期;D:半开期;E:盛开期;***代表C、D、E的表达量相对于A,差异极显著(P<0.001);ns代表没有显著区别;内参基因:18S rRNA;n=3
Fig. 8 Expressions of CnbHLH79 at different flowering stages A: Young bud; B: early bud; C: chromogenic stages of flowering; D: the half-opening stage; E: blooming stage. *** indicates the expressions of C, D, and E shows extremely significant difference compared with that of A(P<0.001). ns means no significant difference. Reference gene: 18S rRNA; n=3
| [1] | 柴胜丰, 韦霄, 蒋运生, 等. 濒危植物金花茶开花物候和生殖构件特征[J]. 热带亚热带植物学报, 2009, 17(1): 5-11. |
| Chai SF, Wei X, Jiang YS, et al. The flowering phenology and characteristics of reproductive modules of endangered plant Camellia nitidissima[J]. J Trop Subtrop Bot, 2009, 17(1): 5-11. | |
| [2] | 陈银霞, 唐山, 赵松子. 金花茶花色遗传研究进展[J]. 南方林业科学, 2015, 43(6): 39-41, 55. |
| Chen YX, Tang S, Zhao SZ. Recent advances in flower color genetics of Camellia nitidissima[J]. South China For Sci, 2015, 43(6): 39-41, 55. | |
| [3] |
Peng X, Yu DY, Feng BM, et al. A new acylated flavonoid glycoside from the flowers of Camellia nitidissima and its effect on the induction of apoptosis in human lymphoma U937 cells[J]. J Asian Nat Prod Res, 2012, 14(8): 799-804.
doi: 10.1080/10286020.2012.691475 URL |
| [4] |
Zhou XW, Li JY, Zhu YL, et al. De novo assembly of the Camellia nitidissima transcriptome reveals key genes of flower pigment biosynthesis[J]. Front Plant Sci, 2017, 8: 1545.
doi: 10.3389/fpls.2017.01545 URL |
| [5] |
Morita Y, Hoshino A. Recent advances in flower color variation and patterning of Japanese morning glory and petunia[J]. Breed Sci, 2018, 68(1): 128-138.
doi: 10.1270/jsbbs.17107 URL |
| [6] |
Wu MJ, Lyu XL, Zhou YJ, et al. High anthocyanin accumulation in an Arabidopsis mutant defective in chloroplast biogenesis[J]. Plant Growth Regul, 2019, 87(3): 433-444.
doi: 10.1007/s10725-019-00481-7 |
| [7] |
Jiang LN, Fan ZQ, Tong R, et al. Functional diversification of the dihydroflavonol 4-reductase from Camellia nitidissima Chi. in the control of polyphenol biosynthesis[J]. Genes, 2020, 11(11): 1341.
doi: 10.3390/genes11111341 URL |
| [8] |
Jiang LN, Fan ZQ, Tong R, et al. Flavonoid 3'-hydroxylase of Camellia nitidissima Chi. promotes the synthesis of polyphenols better than flavonoids[J]. Mol Biol Rep, 2021, 48(5): 3903-3912.
doi: 10.1007/s11033-021-06345-6 |
| [9] |
Zhou XW, Fan ZQ, Chen Y, et al. Functional analyses of a flavonol synthase-like gene from Camellia nitidissima reveal its roles in flavonoid metabolism during floral pigmentation[J]. J Biosci, 2013, 38(3): 593-604.
doi: 10.1007/s12038-013-9339-2 URL |
| [10] | 周兴文, 李纪元, 范正琪. 金花茶查尔酮合成酶基因全长克隆与序列分析[J]. 生物技术通报, 2011(6): 58-64. |
| Zhou XW, Li JY, Fan ZQ. Cloning and sequence analysis of Chalcone synthase gene cDNA from Camellia nitidissima[J]. Biotechnol Bull, 2011(6): 58-64. | |
| [11] |
周兴文, 李纪元, 朱宇林. 金花茶查尔酮合成酶基因CnCHS的克隆及遗传转化研究[J]. 植物研究, 2015, 35(3): 327-332.
doi: 10.7525/j.issn.1673-5102.2015.03.002 |
| Zhou XW, Li JY, Zhu YL. Cloning and genetic transformation of CnCHS gene from Camellia nitidissima[J]. Bull Bot Res, 2015, 35(3): 327-332. | |
| [12] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [13] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [14] |
Zhong CM, Tang Y, Pang B, et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida[J]. Hortic Res, 2020, 7: 78.
doi: 10.1038/s41438-020-0296-2 |
| [15] |
Sun BM, Zhu ZS, Cao PR, et al. Purple foliage coloration in tea(Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1[J]. Sci Rep, 2016, 6: 32534.
doi: 10.1038/srep32534 |
| [16] |
Albert NW, Davies KM, Lewis DH, et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots[J]. Plant Cell, 2014, 26(3): 962-980.
doi: 10.1105/tpc.113.122069 URL |
| [17] |
Zhao AJ, Cui Z, Li TG, et al. mRNA and miRNA expression analysis reveal the regulation for flower spot patterning in Phalaenopsis ‘Panda’[J]. Int J Mol Sci, 2019, 20(17): 4250.
doi: 10.3390/ijms20174250 URL |
| [18] |
Yamagishi M. Isolation and identification of MYB transcription factors(MYB19Long and MYB19Short)involved in raised spot anthocyanin pigmentation in lilies(Lilium spp.)[J]. J Plant Physiol, 2020, 250: 153164.
doi: 10.1016/j.jplph.2020.153164 URL |
| [19] |
Li YQ, Shan XT, Tong LN, et al. The conserved and particular roles of the R2R3-MYB regulator FhPAP1 from Freesia hybrida in flower anthocyanin biosynthesis[J]. Plant Cell Physiol, 2020, 61(7): 1365-1380.
doi: 10.1093/pcp/pcaa065 URL |
| [20] |
郑清冬, 王艺, 欧悦, 等. 兰科植物花色相关基因研究进展[J]. 园艺学报, 2021, 48(10): 2057-2072.
doi: 10.16420/j.issn.0513-353x.2021-0444 |
|
Zheng QD, Wang Y, Ou Y, et al. Research advances of genes responsible for flower colors in Orchidaceae[J]. Acta Hortic Sin, 2021, 48(10): 2057-2072.
doi: 10.16420/j.issn.0513-353x.2021-0444 |
|
| [21] | 王玉书, 杨旭妍, 付震, 等. 观赏羽衣甘蓝BoDFR基因的克隆及表达分析[J]. 西北植物学报, 2020, 40(9): 1483-1489. |
| Wang YS, Yang XY, Fu Z, et al. Cloning and expression analysis of DFR in ornamental kale(Brassica oleracea L. var. acephala)[J]. Acta Bot Boreali Occidentalia Sin, 2020, 40(9): 1483-1489. | |
| [22] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [23] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1): 94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [24] |
Hao YQ, Zong XM, Ren P, et al. Basic helix-loop-helix(bHLH)transcription factors regulate a wide range of functions in Arabidopsis[J]. Int J Mol Sci, 2021, 22(13): 7152.
doi: 10.3390/ijms22137152 URL |
| [25] |
Matus JT, Poupin MJ, Cañón P, et al. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine(Vitis vinifera L.)[J]. Plant Mol Biol, 2010, 72(6): 607-620.
doi: 10.1007/s11103-010-9597-4 pmid: 20112051 |
| [26] |
Kim DH, Park S, Lee JY, et al. Enhancing flower color through simultaneous expression of the B-Peru and mPAP1 transcription factors under control of a flower-specific promoter[J]. Int J Mol Sci, 2018, 19(1): 309.
doi: 10.3390/ijms19010309 URL |
| [27] | Deng CY, Wang JY, Lu CF, et al. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower[J]. Plant Physiol Biochem, 2020, 151: 271-283. |
| [28] |
Li CH, Qiu J, Ding L, et al. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals[J]. Plant Physiol Biochem, 2017, 112: 335-345.
doi: 10.1016/j.plaphy.2017.01.019 URL |
| [29] |
Liu HX, Liu Q, Chen YL, et al. Full-length transcriptome sequencing provides insights into flavonoid biosynthesis in Camellia nitidissima Petals[J]. Gene, 2023, 850: 146924.
doi: 10.1016/j.gene.2022.146924 URL |
| [30] | 李辛雷, 王佳童, 孙振元, 等. 五种金花茶组植物类黄酮成分及其与花色关系[J]. 生态学杂志, 2019, 38(4): 961-966. |
| Li XL, Wang JT, Sun ZY, et al. Flavonoid components and their relationship with flower colors in five species of Camellia section Chrysantha[J]. Chin J Ecol, 2019, 38(4): 961-966. | |
| [31] | 姜丽娜, 李纪元, 童冉, 等. 金花茶组植物花色与细胞内重要环境因子的关系[J]. 广西植物, 2019, 39(12): 1605-1612. |
| Jiang LN, Li JY, Tong R, et al. Relationship between flower color and important cellular environment elemental factors in yellow Camellia[J]. Guihaia, 2019, 39(12): 1605-1612. | |
| [32] |
Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology[J]. Plant Physiol, 2001, 126(2): 485-493.
doi: 10.1104/pp.126.2.485 pmid: 11402179 |
| [33] |
Agati G, Tattini M. Multiple functional roles of flavonoids in photoprotection[J]. New Phytol, 2010, 186(4): 786-793.
doi: 10.1111/j.1469-8137.2010.03269.x pmid: 20569414 |
| [34] | Jaakola L, Hohtola A. Effect of latitude on flavonoid biosynthesis in plants[J]. Plant Cell Environ, 2010, 33(8): 1239-1247. |
| [35] |
An JP, Li R, Qu FJ, et al. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple[J]. Plant J, 2018, 96(3): 562-577.
doi: 10.1111/tpj.14050 URL |
| [36] |
Samarina LS, Bobrovskikh AV, Doroshkov AV, et al. Comparative expression analysis of stress-inducible candidate genes in response to cold and drought in tea plant[Camellia sinensis(L.) kuntze][J]. Front Genet, 2020, 11: 611283.
doi: 10.3389/fgene.2020.611283 URL |
| [37] |
Wang ZY, Zhang YM, Hu HF, et al. CabHLH79 acts upstream of CaNAC035 to regulate cold stress in pepper[J]. Int J Mol Sci, 2022, 23(5): 2537.
doi: 10.3390/ijms23052537 URL |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [3] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [4] | 杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156. |
| [5] | 镐青青, 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶. 马尾松NAC转录因子基因PmNAC8的克隆及表达分析[J]. 生物技术通报, 2022, 38(4): 202-216. |
| [6] | 赵婷婷, 王俊刚, 王文治, 冯翠莲, 冯小艳, 张树珍. 甘蔗单糖转运蛋白基因ShSTP7序列分析及组织表达特征测定[J]. 生物技术通报, 2022, 38(4): 72-78. |
| [7] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| [8] | 骆鹰, 谭智, 王帆, 刘晓霞, 罗小芳, 何福林. 银杏GbR2R3-MYB1基因的克隆及非生物胁迫应答分析[J]. 生物技术通报, 2022, 38(10): 184-194. |
| [9] | 孙瑞芬, 张艳芳, 牛素清, 郭树春, 李素萍, 于海峰, 聂惠, 牟英男. 向日葵HaACO1基因的表达分析及功能验证[J]. 生物技术通报, 2021, 37(9): 114-124. |
| [10] | 范亚朋, 芮存, 张悦新, 陈修贵, 陆许可, 王帅, 张红, 徐楠, 王晶, 陈超, 叶武威. 陆地棉耐碱基因GHZAT12的克隆、表达及生物信息学分析[J]. 生物技术通报, 2021, 37(8): 121-130. |
| [11] | 山草梅, 叶蕾, 张连虎, 况卫刚, 孙晓棠, 马建, 崔汝强. 水稻抗潜根线虫基因OsRAI1的克隆及功能分析[J]. 生物技术通报, 2021, 37(7): 146-155. |
| [12] | 孙小倩, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子FtMYBF的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2021, 37(3): 10-17. |
| [13] | 韩占红, 宗元元, 张学梅, 王斌, PRUSKY Dov, 毕阳. 扩展青霉erg4的生物信息学、亚细胞定位及表达分析[J]. 生物技术通报, 2021, 37(12): 60-70. |
| [14] | 张睿珠, 江羽宸, 黄俊, 闫洁. 橡胶草SRPP2基因克隆及表达分析[J]. 生物技术通报, 2020, 36(1): 9-14. |
| [15] | 段敏杰, 伊洪伟, 王进, 武峥. 梨黑斑病抗病相关基因PpEMS1的克隆与分析[J]. 生物技术通报, 2019, 35(11): 16-21. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||