








生物技术通报 ›› 2023, Vol. 39 ›› Issue (6): 259-273.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1316
杨洋1( ), 朱金成1, 娄慧1, 韩泽刚2(
), 朱金成1, 娄慧1, 韩泽刚2( ), 张薇1(
), 张薇1( )
)
收稿日期:2022-10-26
出版日期:2023-06-26
发布日期:2023-07-07
通讯作者:
张薇,女,博士,教授,研究方向:棉花分子育种;E-mail: zhw_agr@shzu.edu.cn;作者简介:杨洋,女,硕士研究生,研究方向:棉花分子育种;E-mail: 1977277872@qq.com
基金资助:
YANG Yang1( ), ZHU Jin-cheng1, LOU Hui1, HAN Ze-gang2(
), ZHU Jin-cheng1, LOU Hui1, HAN Ze-gang2( ), ZHANG Wei1(
), ZHANG Wei1( )
)
Received:2022-10-26
Published:2023-06-26
Online:2023-07-07
摘要:
棉花枯萎病是棉花生产中常见的一种病害,目前在海岛棉中发病严重,直接影响海岛棉产量和品质,对海岛棉产业发展造成巨大威胁。为解析棉花与枯萎病菌尖孢镰刀菌互作的分子机制,以尖孢镰刀菌侵染48 h的抗感棉花根部组织及致病菌体为材料,利用RNA-seq测序技术分析尖孢镰刀菌与棉花互作的基因表达特性。结果表明,在抗感棉花品种中分别检测到15 218和9 358个差异基因,在侵染抗感棉花品种的尖孢镰刀菌中分别检测到3 708和3 656个差异基因。GO功能富集分析发现,互作后棉花中主要为氧化应激反应、生长素-激活信号通路、对刺激的反应、对受伤的反应和转录因子活性等基因功能;KEGG显著富集到内吞作用、植物激素信号转导、氨基酸生物合成、碳代谢、植物-病原菌互作、苯丙烷类生物合成等代谢途径,抗病品种在调控对刺激的反应和对受伤的反应中上调显著。GO功能富集分析发现,互作后尖孢镰刀菌中的差异表达基因多参与膜的组成部分、催化活性调节、ATP结合等类别;KEGG显著富集到过氧化物酶体,丝裂原活化蛋白激酶信号通路,缬氨酸、亮氨酸和异亮氨酸降解,甘氨酸、丝氨酸和苏氨酸代谢,碳代谢,氨基酸生物合成,淀粉和蔗糖代谢等通路。本研究为棉花响应枯萎病胁迫以及尖孢镰刀菌致病性研究提供了丰富的基因资源,为深入解析尖孢镰刀菌与棉花互作的机制研究奠定基础。
杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273.
YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum[J]. Biotechnology Bulletin, 2023, 39(6): 259-273.
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| Gb2763 | TTCCACAGTTCCAGGGTA | GATGATCCAAGGCTTTCT |
| Gb1474 | AAACAAGGTAAGGAGGCT | GTTGGGTAAATAATAGGC |
| Gb7593 | CAACTTGGCATCTTATCC | CTTACATTGAGCAGCAAC |
| Gb8190 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb8191 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb9261 | TTTGCCAAAGAGCGTAGA | TTAAAGAATGCGGTGTCG |
| Gb9263 | TTTCGGCAAGTCTATGAT | TCTTTCTGGTCGTGGATG |
| FOTG10073 | GATGCCTGCCGTGGTGGAT | GGGCGAAGAAACAGTGTAAAGC |
| FOTG05109 | TATCGCCCAGATGTTCAA | TACCACGGTGCTGTTCCA |
| FOTG15482 | TTTCTTCATCGCCCTGTA | ACATTGCCGACTTGGATT |
| FOTG11725 | TTGTTGGTTTGCCCTTGT | CGAGCGGTCTGAGTGTAG |
| FOTG17884 | GCAATCTCAGCCTCAAGTT | GACAAGCGTTCATCCATAA |
| FOTG17663 | GCCTTCTCGGATCTTCTA | GCAATGCTGGGATGGTAT |
| GhUBQ7 | GAAGACCTACACCAAGCCCAA | CGGACTCTACTCAATCCCCACC |
表1 RT-qPCR所用的引物
Table 1 Primers for RT-qPCR
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| Gb2763 | TTCCACAGTTCCAGGGTA | GATGATCCAAGGCTTTCT |
| Gb1474 | AAACAAGGTAAGGAGGCT | GTTGGGTAAATAATAGGC |
| Gb7593 | CAACTTGGCATCTTATCC | CTTACATTGAGCAGCAAC |
| Gb8190 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb8191 | CATGGAGCAAAGGTTTAA | TGTTGGATTCAGGGAGAT |
| Gb9261 | TTTGCCAAAGAGCGTAGA | TTAAAGAATGCGGTGTCG |
| Gb9263 | TTTCGGCAAGTCTATGAT | TCTTTCTGGTCGTGGATG |
| FOTG10073 | GATGCCTGCCGTGGTGGAT | GGGCGAAGAAACAGTGTAAAGC |
| FOTG05109 | TATCGCCCAGATGTTCAA | TACCACGGTGCTGTTCCA |
| FOTG15482 | TTTCTTCATCGCCCTGTA | ACATTGCCGACTTGGATT |
| FOTG11725 | TTGTTGGTTTGCCCTTGT | CGAGCGGTCTGAGTGTAG |
| FOTG17884 | GCAATCTCAGCCTCAAGTT | GACAAGCGTTCATCCATAA |
| FOTG17663 | GCCTTCTCGGATCTTCTA | GCAATGCTGGGATGGTAT |
| GhUBQ7 | GAAGACCTACACCAAGCCCAA | CGGACTCTACTCAATCCCCACC |
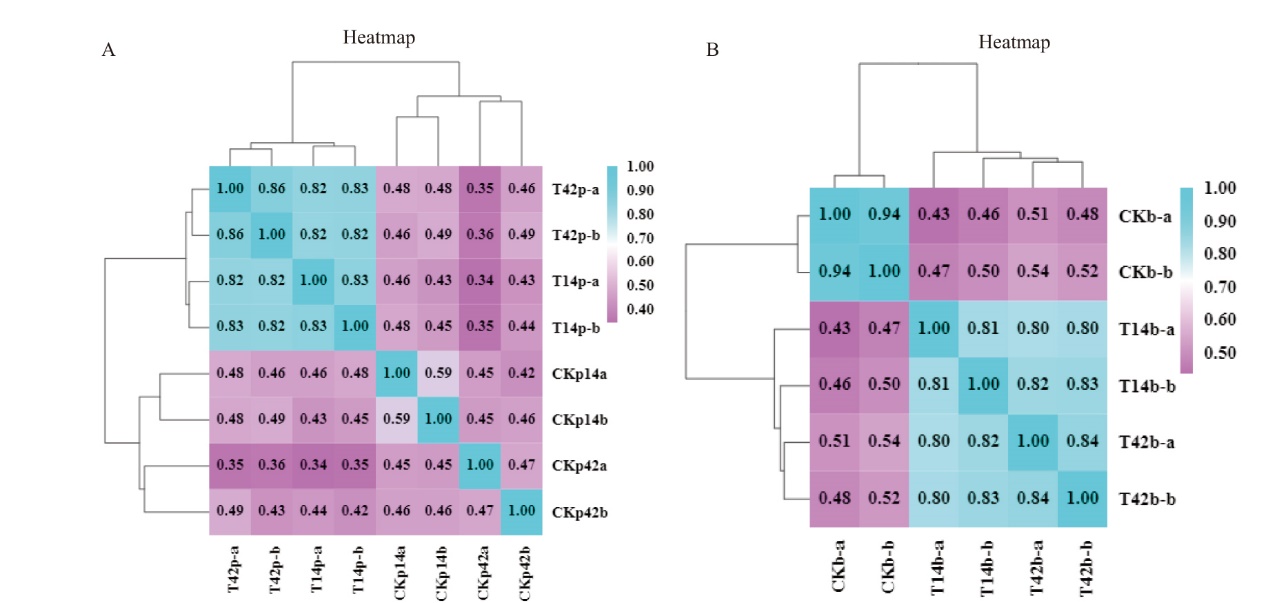
图1 样品间相关性检验 A:4个棉花根系组织样品的相关性检验,其中T14p为新海14号实验组样品,T42p为新海42号实验组样品,CKp14为新海14号对照组样品,CKp42为新海42号对照组样品;B:3个枯萎病菌样品的相关性检验,CKb为枯萎病菌对照组样品,T14b为新海14号枯萎病菌样品,T42b为新海42号枯萎病菌样品;a和b为两个重复
Fig. 1 Correlation analysis among samples A: Correlation analysis of four samples from cotton root tissues. T14p indicates the treatment sample from Xinhai 14, T42p indicates the treatment sample from Xinhai 42, CKp14 indicates the control sample from Xinhai 14 and CKp42 indicates the control sample from Xinhai 42. B: Correlation analysis of three samples from Fov. CKb refers to the control sample of Fov, T14b to the Fov sample obtained from Xinhai 14, T42b to the Fov sample obtained from Xinhai 42. a and b are the duplications
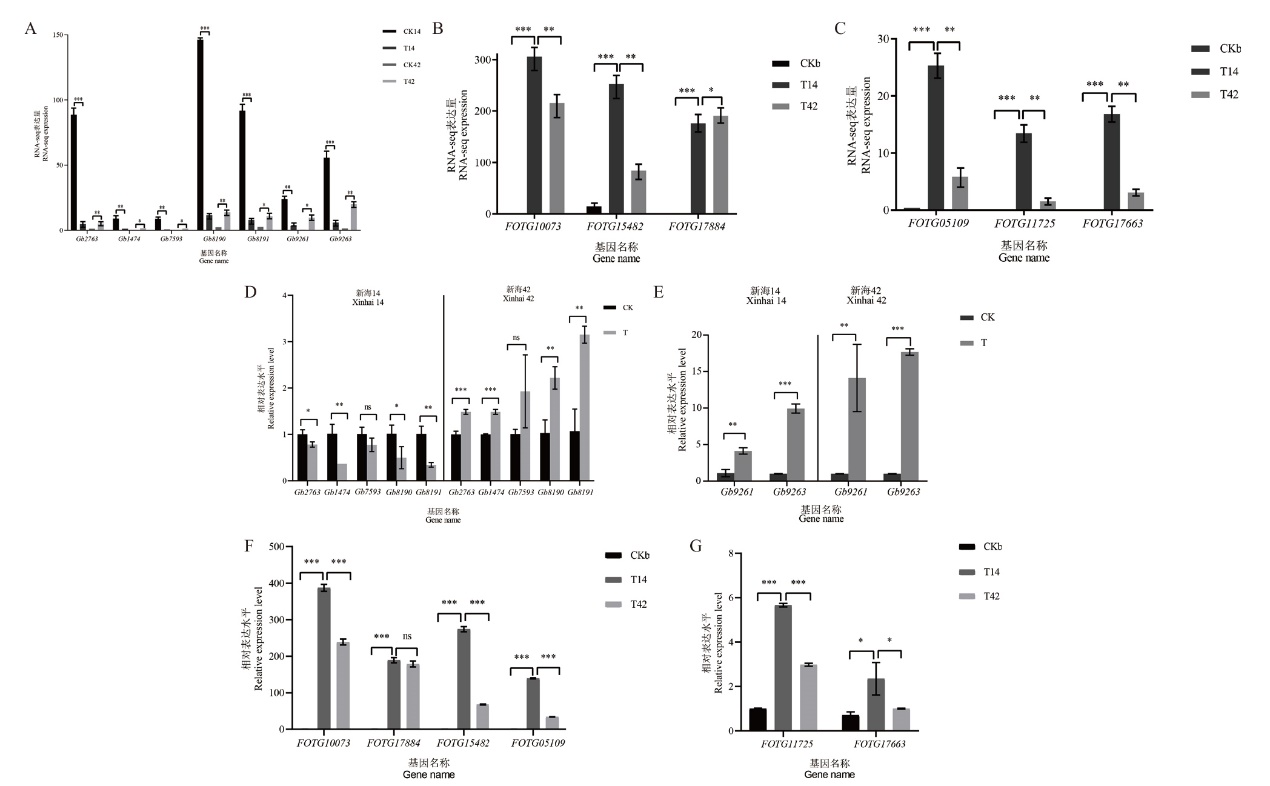
图2 RT-qPCR 验证RNA-seq 结果 A-C:RNA-seq表达量图;D和E:棉花中7个基因的表达模式;F和G:尖孢镰刀菌中6个基因的表达模式;CK为对照组;T为实验组;CKb为菌对照,T14为新海14实验组,T42为新海42号实验组。*表示有差异(P<0.05),**表示差异显著(P<0.01),***表示差异极显著(P<0.001)
Fig. 2 Validation of RNA-seq data by RT-qPCR A-C : RNA-seq expression. D and E : RT-qPCR verification of 7 genes in cotton. F and G : RT-qPCR verification of 6 genes in Fusarium oxysporum. CK: Control group. T: Treatment group. CKb: Bacteria control group. T14: Xinhai 14 treatment group.T42: Xinhai 42 treatment group. * indicates difference(P<0.05), ** indicates significant difference(P<0.01), and *** indicates extremely significant difference(P<0.001)
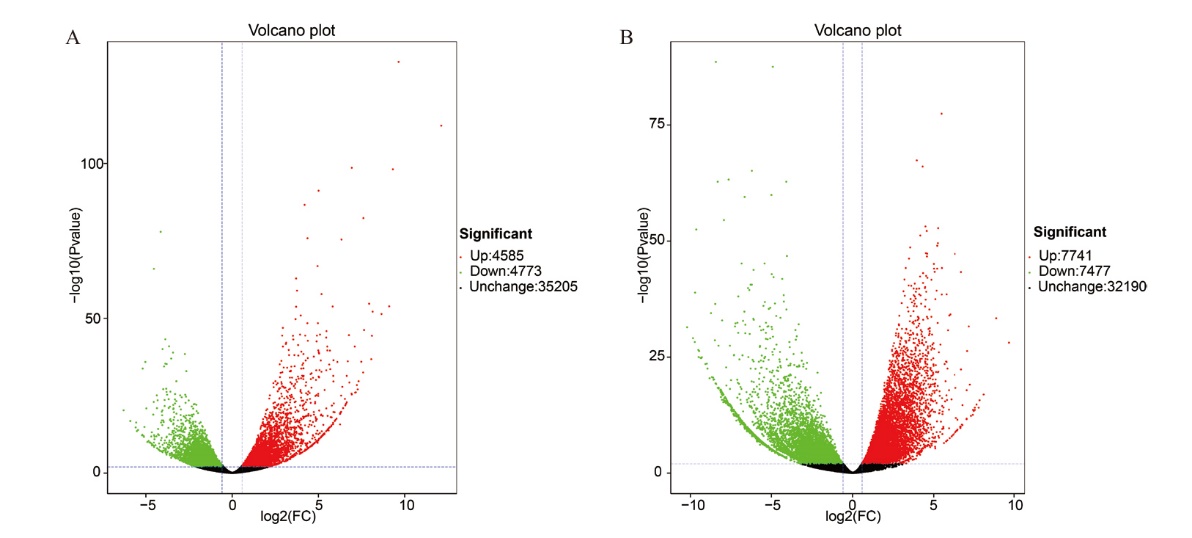
图3 不同组间棉花差异表达基因火山图 A:CK14_T14;B:CK42_T42。图中绿色为下调基因,红色为上调基因。下同
Fig. 3 Volcano map of differentially expressed genes in the cotton of different treatments Green is down-regulated gene and red is up- regulated gene. The same below

图5 新海14接菌后差异表达基因GO分析 A-C:上调基因的GO分析;D-F:下调基因的GO分析。横坐标为每个条目富集到的基因数,纵坐标为GO条目
Fig. 5 GO analysis of differentially expressed genes in Xinhai 14 after inoculation A-C: GO analysis of up-regulated genes. D-F: GO analysis of down-regulated genes. The abscissa is number of genes enriched for each term, and the ordinate is the term of GO level
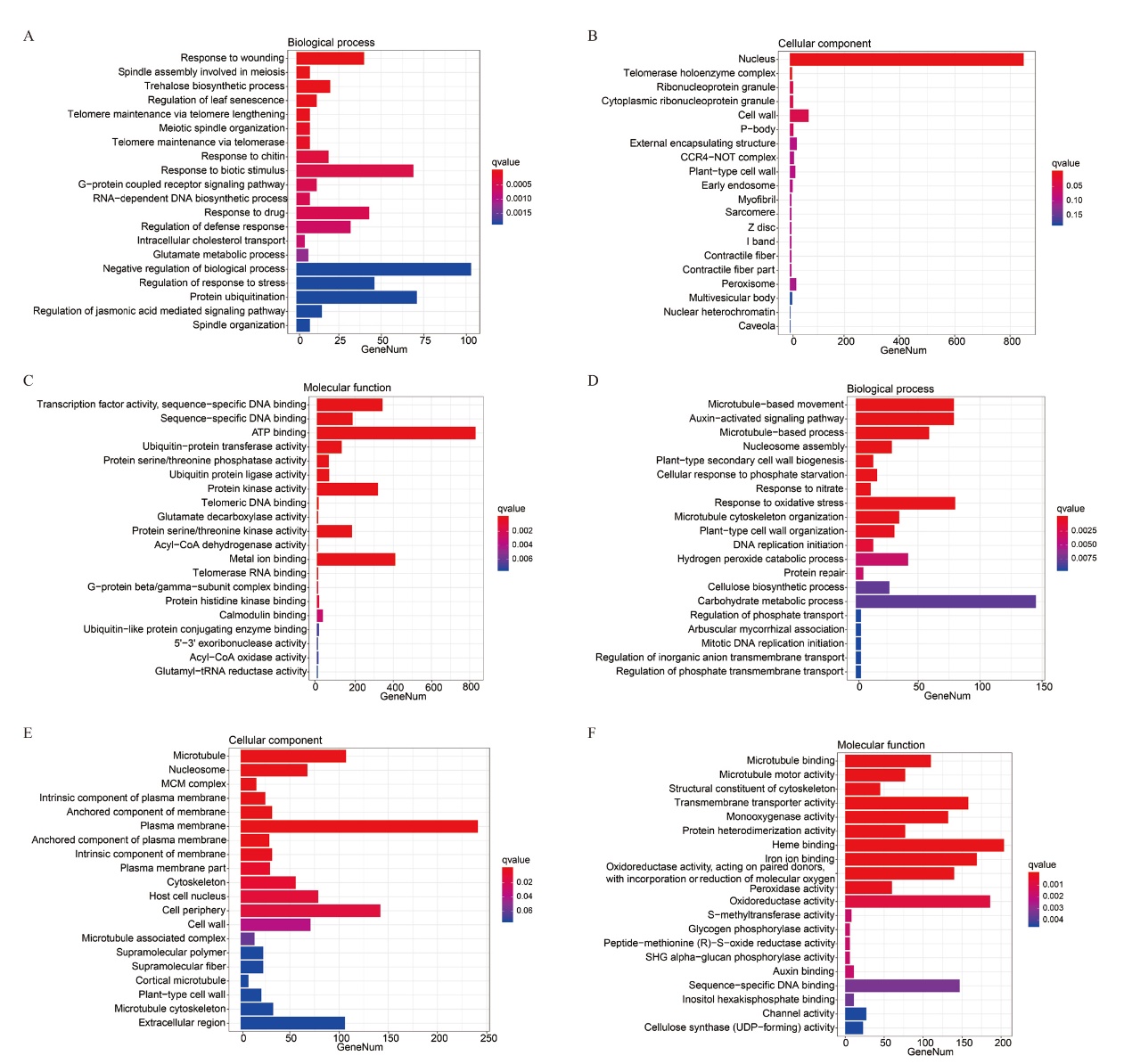
图6 新海42接菌后差异表达基因GO分析 A-C:上调基因的GO分析;D-F:下调基因的GO分析。横坐标为每个条目富集到的基因数,纵坐标为GO条目
Fig. 6 GO analysis of differentially expressed genes in Xinhai 42 after inoculation A-C: GO analysis of up-regulated genes. D-F: GO analysis of down-regulated genes. The abscissa is number of genes enriched for each term,the ordinate is the term of GO level
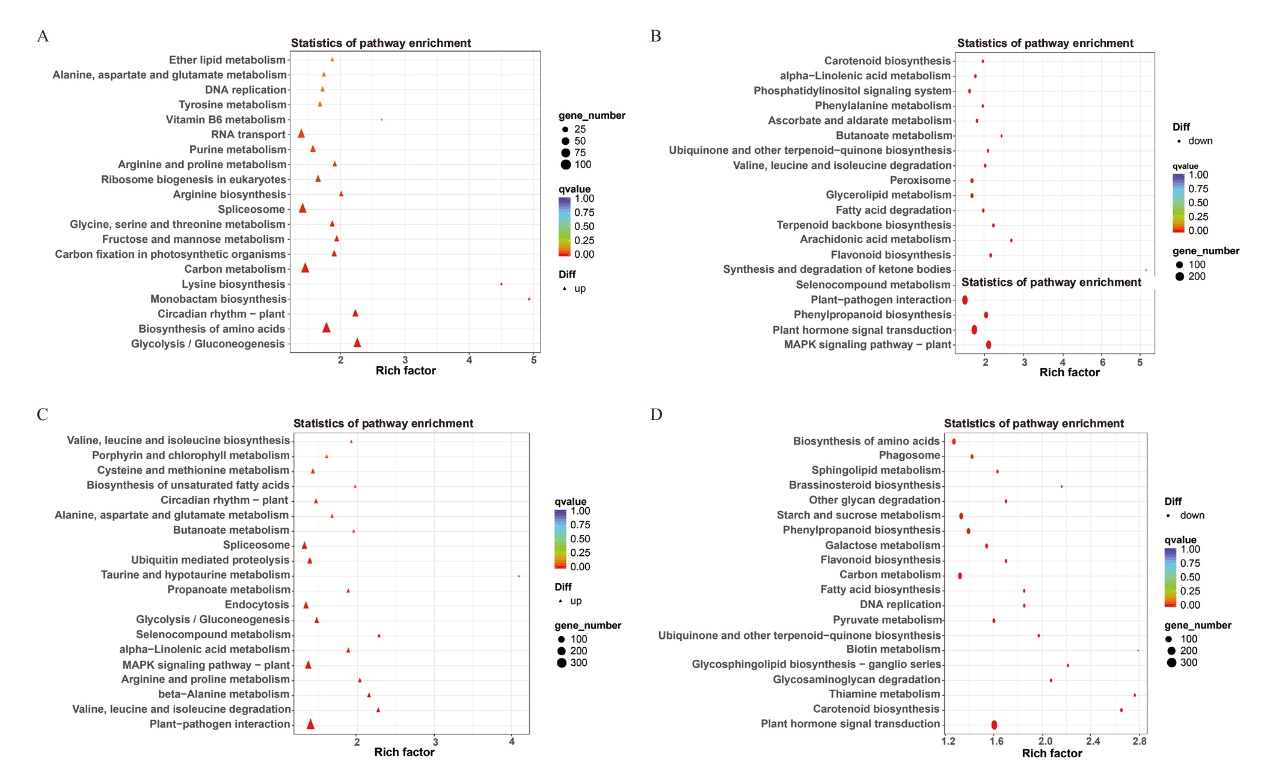
图7 接菌后两个棉花品种中差异表达基因KEGG分析 A:新海14号上调基因的KEGG分析;B:新海14号下调基因的KEGG分析;C:新海42号上调基因的KEGG分析;D:新海42号下调基因的KEGG分析
Fig. 7 KEGG analysis of differentially expressed genes in two cotton cultivars after inoculation A: KEGG analysis of up-regulated genes in Xinhai 14. B: KEGG analysis of down-regulated genes in Xinhai 14. C: KEGG analysis of up-regulated genes in Xinhai 42. D: KEGG analysis of down-regulated genes in Xinhai 42

图8 尖孢镰刀菌侵染新海14后差异表达基因GO分析 A-C:新海14号尖孢镰刀菌中上调基因的GO分析;D-F:新海14号尖孢镰刀菌中下调基因的GO分析。横坐标为每个条目富集到的基因数,纵坐标为GO条目
Fig. 8 GO analysis of differentially expressed genes of Fov after infecting Xinhai 14 A-C: GO analysis of up-regulated genes of Fov in Xinhai 14. D-F: GO analysis of down-regulated genes of Fov in Xinhai 14. The abscissa is number of genes enriched for each term, the ordinate is the term of GO level
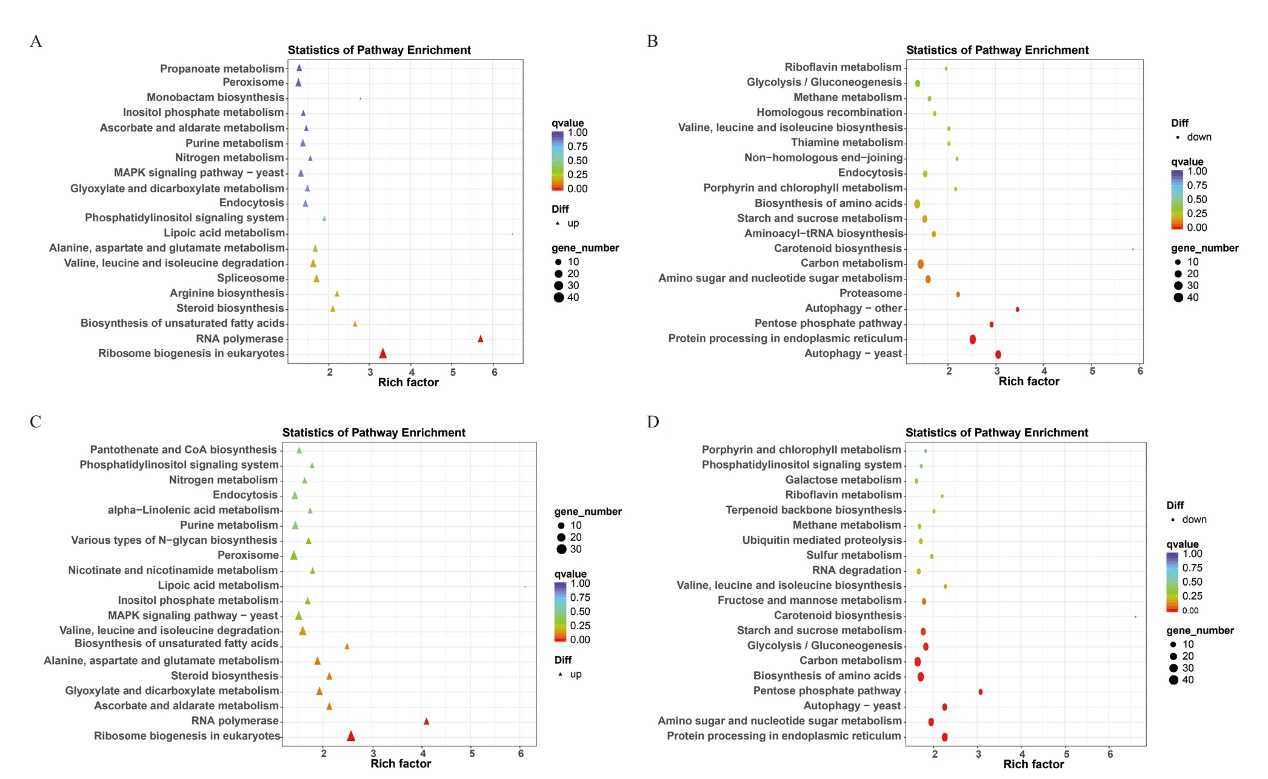
图9 尖孢镰刀菌侵染新海14和新海42后差异表达基因KEGG分析 A:新海14号中尖孢镰刀菌上调基因的KEGG分析;B:新海14号中尖孢镰刀菌下调基因的KEGG分析;C:新海42号中尖孢镰刀菌上调基因的KEGG分析;D:新海42号中尖孢镰刀菌下调基因的KEGG分析
Fig. 9 KEGG analysis of differentially expressed genes of Fov after infecting Xinhai 14 and Xinhai 42 A: KEGG analysis of up-regulated genes of Fov(Fusarium oxysporum f. sp. Vasinfectum)in Xinhai 14. B: KEGG analysis of down-regulated genes of Fov in Xinhai 14. C: KEGG analysis of up-regulated genes of Fov in Xinhai 42. D: KEGG analysis of down-regulated genes of Fov in Xinhai 42

图10 尖孢镰刀菌侵染新海42后差异表达基因GO分析 A-C:新海42号尖孢镰刀菌中上调基因的GO分析;D-F:新海42号尖孢镰刀菌中下调基因的GO分析。横坐标为每个条目富集到的基因数,纵坐标为GO条目
Fig. 10 GO analysis of differentially expressed genes of Fov after infecting Xinhai 42 A-C: GO analysis of up-regulated genes of Fov in Xinhai 42. D-F: GO analysis of down-regulated genes of Fov in Xinhai 42. The abscissa is number of genes enriched for each term, the ordinate is the term of GO level
| [16] | 雷忠华. 基于互作转录组分析的抗病香蕉品系南天蕉与尖孢镰刀菌互作机理研究[D]. 广州: 华南理工大学, 2018. |
| Lei ZH. Dual RNA-sequencing unveils host-pathogen interactions in resistant banana cultivars Nantianjiao upon Fusarium oxysporum race 4 challenge[D]. Guangzhou: South China University of Technology, 2018. | |
| [17] |
Ma ZY, Zhang Y, Wu LQ, et al. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement[J]. Nat Genet, 2021, 53(9): 1385-1391.
doi: 10.1038/s41588-021-00910-2 pmid: 34373642 |
| [18] |
刘戈辉, 韩泽刚, 孙士超, 等. 转GhB301基因棉花响应枯萎病菌侵染的转录组分析[J]. 核农学报, 2021, 35(12): 2733-2745.
doi: 10.11869/j.issn.100-8551.2021.12.2733 |
| Liu GH, Han ZG, Sun SC, et al. Transcriptome analysis of GhB301 transgenic cotton response to Fusarium wilt infection[J]. J Nucl Agric Sci, 2021, 35(12): 2733-2745. | |
| [19] |
Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias[J]. Genome Biol, 2010, 11(2): R14.
doi: 10.1186/gb-2010-11-2-r14 URL |
| [20] |
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Res, 2000, 28(1): 27-30.
doi: 10.1093/nar/28.1.27 pmid: 10592173 |
| [21] |
Spoel SH, Dong XN. How do plants achieve immunity? Defence without specialized immune cells[J]. Nat Rev Immunol, 2012, 12(2): 89-100.
doi: 10.1038/nri3141 pmid: 22273771 |
| [22] |
Xiong DG, Wang YL, Ma J, et al. Deep mRNA sequencing reveals stage-specific transcriptome alterations during microsclerotia development in the smoke tree vascular wilt pathogen, Verticillium dahliae[J]. BMC Genomics, 2014, 15(1): 324.
doi: 10.1186/1471-2164-15-324 |
| [23] |
Berens ML, Berry HM, Mine A, et al. Evolution of hormone signaling networks in plant defense[J]. Annu Rev Phytopathol, 2017, 55: 401-425.
doi: 10.1146/annurev-phyto-080516-035544 pmid: 28645231 |
| [24] |
Bürger M, Chory J. Stressed out about hormones: how plants orchestrate immunity[J]. Cell Host Microbe, 2019, 26(2): 163-172.
doi: S1931-3128(19)30350-6 pmid: 31415749 |
| [1] |
Han ZG, Chen H, Cao YW, et al. Genomic insights into genetic improvement of upland cotton in the world's largest growing region[J]. Ind Crops Prod, 2022, 183: 114929.
doi: 10.1016/j.indcrop.2022.114929 URL |
| [2] | 朱永军, 张西英, 李金荣, 等. 海岛棉枯萎病抗性遗传规律及分子标记的初步筛选[J]. 新疆农业科学, 2010, 47(2): 268-273. |
| Zhu YJ, Zhang XY, Li JR, et al. Inheritance of resistance to Fusarium wilt and their molecular marker in Gossypium barbadense[J]. Xinjiang Agric Sci, 2010, 47(2): 268-273. | |
| [3] |
Michielse CB, Rep M. Pathogen profile update: Fusarium oxysporum[J]. Mol Plant Pathol, 2009, 10(3): 311-324.
doi: 10.1111/j.1364-3703.2009.00538.x pmid: 19400835 |
| [4] |
Back MA, Haydock PPJ, Jenkinson P. Disease complexes involving plant parasitic nematodes and soilborne pathogens[J]. Plant Pathol, 2002, 51(6): 683-697.
doi: 10.1046/j.1365-3059.2002.00785.x URL |
| [5] |
Wang C, Roberts PA. A Fusarium Wilt resistance gene in Gossypium barbadense and its effect on root-knot nematode-wilt disease complex[J]. Phytopathology, 2006, 96(7): 727-734.
doi: 10.1094/PHYTO-96-0727 pmid: 18943146 |
| [6] | 黄启秀. 海岛棉类黄酮代谢途径中与枯萎病抗性相关基因的克隆及功能验证[D]. 乌鲁木齐: 新疆农业大学, 2017. |
| Huang QX. Cloning and functional verification of genes related to Fusarium wilt resistance in flavonoid metabolism pathway of island cotton(Gossypium barbadense L.)[D]. Urumqi: Xinjiang Agricultural University, 2017. | |
| [7] | 赵曾强. 棉花GhWRKY44和GhWRKY22基因的克隆及功能初步分析[D]. 石河子: 石河子大学, 2015. |
| Zhao ZQ. Cloning and functional analysis of GhWRKY44 and GhWRKY22 gene in cotton[D]. Shihezi: Shihezi University, 2015. | |
| [8] | 张艳艳, 季苇芹, 杨玉文, 等. 转录组学技术及其在瓜菜作物上的应用[J]. 中国瓜菜, 2016, 29(12): 1-5, 13. |
| Zhang YY, Ji WQ, Yang YW, et al. The technology of transcriptome and its application in cucurbit and vegetable crops[J]. China Cucurbits Veg, 2016, 29(12): 1-5, 13. | |
| [9] |
Zhang WW, Zhang HC, Liu K, et al. Large-scale identification of Gossypium hirsutum genes associated with Verticillium dahliae by comparative transcriptomic and reverse genetics analysis[J]. PLoS One, 2017, 12(8): e0181609.
doi: 10.1371/journal.pone.0181609 URL |
| [10] |
Xu L, Zhu LF, Tu LL, et al. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry[J]. J Exp Bot, 2011, 62(15): 5607-5621.
doi: 10.1093/jxb/err245 pmid: 21862479 |
| [11] |
Jun Z, Zhang ZY, Gao YL, et al. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt[J]. Sci Rep, 2015, 5: 15048.
doi: 10.1038/srep15048 |
| [12] |
Zhang LS, Ni H, Du X, et al. The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections[J]. New Phytol, 2017, 215(1): 368-381.
doi: 10.1111/nph.2017.215.issue-1 URL |
| [13] | 冯志迪. V型肌球蛋白在大丽轮枝菌与棉花互作过程中的机制研究[D]. 石河子: 石河子大学, 2017. |
| Feng ZD. Essential roles of the myosin V in plant cotton-verticillum interaction[D]. Shihezi: Shihezi University, 2017. | |
| [14] |
Soanes DM, Chakrabarti A, Paszkiewicz KH, et al. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae[J]. PLoS Pathog, 2012, 8(2): e1002514.
doi: 10.1371/journal.ppat.1002514 URL |
| [15] | 李成伟. 番茄与白粉菌互作的细胞学和转录组学分析[D]. 北京: 中国农业科学院, 2007. |
| Li CW. The cytological and transcriptomic investigations into interactions of tomato and powdery mildew[D]. Beijing: Chinese Academy of Agricultural Sciences, 2007. | |
| [25] |
Duressa D, Anchieta A, Chen DQ, et al. RNA-seq analyses of gene expression in the microsclerotia of Verticillium dahliae[J]. BMC Genomics, 2013, 14: 607.
doi: 10.1186/1471-2164-14-607 |
| [26] |
Zou Q, Luo S, Wu HT, et al. A GMC oxidoreductase GmcA is required for symbiotic nitrogen fixation in Rhizobium leguminosarum bv. viciae[J]. Front Microbiol, 2020, 11: 394.
doi: 10.3389/fmicb.2020.00394 URL |
| [27] | Arun KS, Vinay S, Jyoti S. A review on strain improvement of fungi for enhanced lipase production[J]. Journal of Biology and Nature, 2017, 6(3): 173-180. |
| [28] |
Kim YK, Wang YH, Liu ZM, et al. Identification of a hard surface contact-induced gene in Colletotrichum gloeosporioides conidia as a sterol glycosyl transferase, a novel fungal virulence factor[J]. Plant J, 2002, 30(2): 177-187.
doi: 10.1046/j.1365-313X.2002.01284.x URL |
| [29] |
Shen M, Zhao DK, Qiao Q, et al. Identification of glutathione S-transferase(GST)genes from a dark septate endophytic fungus(Exophiala pisciphila)and their expression patterns under varied metals stress[J]. PLoS One, 2015, 10(4): e0123418.
doi: 10.1371/journal.pone.0123418 URL |
| [1] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [2] | 赵志祥, 王殿东, 周亚林, 王培, 严婉荣, 严蓓, 罗路云, 张卓. 枯草芽孢杆菌Ya-1对辣椒枯萎病的防治及其对根际真菌群落的影响[J]. 生物技术通报, 2023, 39(9): 213-224. |
| [3] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [4] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [5] | 褚睿, 李昭轩, 张学青, 杨东亚, 曹行行, 张雪艳. 黄瓜枯萎病拮抗芽孢杆菌的筛选、鉴定及其生防潜力[J]. 生物技术通报, 2023, 39(8): 262-271. |
| [6] | 赵林艳, 徐武美, 王豪吉, 王昆艳, 魏富刚, 杨绍周, 官会林. 施用生物炭对连作三七根际真菌群落与存活率的影响[J]. 生物技术通报, 2023, 39(7): 219-227. |
| [7] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [8] | 申云鑫, 施竹凤, 周旭东, 李铭刚, 张庆, 冯路遥, 陈齐斌, 杨佩文. 三株具生防功能芽孢杆菌的分离鉴定及其生物活性研究[J]. 生物技术通报, 2023, 39(3): 267-277. |
| [9] | 邓嘉辉, 雷建峰, 赵燚, 刘敏, 胡子曜, 尤扬子, 邵武奎, 柳建飞, 刘晓东. 基于Csy4与MCP的新型迷你基因组编辑系统的构建[J]. 生物技术通报, 2023, 39(10): 68-79. |
| [10] | 朱金成, 杨洋, 娄慧, 张薇. 外源褪黑素调控棉花枯萎病抗性研究[J]. 生物技术通报, 2023, 39(1): 243-252. |
| [11] | 孙卓, 王妍, 韩忠明, 王云贺, 赵淑杰, 杨利民. 防风根际真菌的分离鉴定及其生防潜力评价[J]. 生物技术通报, 2023, 39(1): 264-273. |
| [12] | 李霁虹, 荆玉玲, 马桂珍, 郭荣君, 李世东. 无色杆菌77的基因组构成及其趋化和耐药特性[J]. 生物技术通报, 2022, 38(9): 136-146. |
| [13] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| [14] | 金姣姣, 刘自刚, 米文博, 徐明霞, 邹娅, 徐春梅, 赵彩霞. 利用RNA-Seq鉴定调控甘蓝型油菜叶片光合特性的低温胁迫应答基因[J]. 生物技术通报, 2022, 38(4): 126-142. |
| [15] | 赵曾强, 郭文婷, 张析, 李潇玲, 张薇. 棉花抗枯萎病相关基因GhERF5-4D的克隆及功能分析[J]. 生物技术通报, 2022, 38(4): 193-201. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||