








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 261-268.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0010
张嘉华1( ), 张慧梅1, 马喜喜1, 孙焱森1, 李若冰1, 李柠杏1, 才学鹏2, 乔军1, 孟庆玲1(
), 张慧梅1, 马喜喜1, 孙焱森1, 李若冰1, 李柠杏1, 才学鹏2, 乔军1, 孟庆玲1( )
)
收稿日期:2024-01-04
出版日期:2024-05-26
发布日期:2024-04-19
通讯作者:
孟庆玲,女,博士,教授,研究方向:动物寄生虫学;E-mail: xjmqlqj@163.com作者简介:张嘉华,男,硕士研究生,研究方向:动物寄生虫学;E-mail: zhangjiahua0309@163.com
基金资助:
ZHANG Jia-hua1( ), ZHANG Hui-mei1, MA Xi-xi1, SUN Yan-sen1, LI Ruo-bing1, LI Ning-xing1, CAI Xue-peng2, QIAO Jun1, MENG Qing-ling1(
), ZHANG Hui-mei1, MA Xi-xi1, SUN Yan-sen1, LI Ruo-bing1, LI Ning-xing1, CAI Xue-peng2, QIAO Jun1, MENG Qing-ling1( )
)
Received:2024-01-04
Published:2024-05-26
Online:2024-04-19
摘要:
【目的】为探究捕食线虫真菌少孢节丛孢菌几丁质诱导过程中分泌蛋白AO-492的生物学功能。【方法】对少孢节丛孢菌几丁质酶AO-492主要结构域编码区进行基因克隆及分子特征分析,并在毕赤酵母中进行表达。利用镍柱亲和层析法纯化重组蛋白ReAO-Z492,采用NAG检测法分析了该重组蛋白在不同温度、pH及金属离子条件下的酶学活性,并将其作用于秀丽隐杆线虫及虫卵分析其生物学功能。【结果】几丁质酶AO-492有信号肽,无跨膜结构域,含有两个几丁质结合结构域和一个糖苷水解酶18家族结构域,并含有糖苷水解酶18家族几丁质酶高度保守的底物结合位点-SVGGWT-和水解酶活性位点-FDGGDLDWE-,含有典型的TIM桶形分子结构。系统进化分析显示,该蛋白与坚粘孢单顶孢几丁质酶(EPS35099.1)的亲缘关系相对最近。SDS-PAGE和Western Blot分析表明,ReAO-Z492分子量约为59 kD,可与小鼠抗少孢节丛孢菌多克隆抗体发生特异性反应。ReAO-Z492最适温度为40℃,最适pH为7.0;Mg2+对其酶活有促进作用,而Ag+、Cu2+、Fe3+和Zn2+有抑制作用。ReAO-Z492对秀丽隐杆线虫体壁及其虫卵卵壳有较强的降解活性。【结论】少孢节丛孢菌几丁质酶 AO-492 对秀丽隐杆线虫及虫卵具有较强的降解作用。
张嘉华, 张慧梅, 马喜喜, 孙焱森, 李若冰, 李柠杏, 才学鹏, 乔军, 孟庆玲. 少孢节丛孢菌几丁质酶AO-492对线虫的降解作用研究[J]. 生物技术通报, 2024, 40(5): 261-268.
ZHANG Jia-hua, ZHANG Hui-mei, MA Xi-xi, SUN Yan-sen, LI Ruo-bing, LI Ning-xing, CAI Xue-peng, QIAO Jun, MENG Qing-ling. Degradation of Nematodes by Chitinase AO-492 from Arthrospora oligospora[J]. Biotechnology Bulletin, 2024, 40(5): 261-268.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product size/bp |
|---|---|---|
| 492-F 492-R 492-ZF 492-ZR | ATGAGATCTCTGTTTCTACGAG TCAACAGTCTCTCGGCAGTAAG GCTACGTAATGCATCATCATCATCATCATAGATCTCTGTTTCTACGA GCGGCCGCTCAACAGTCTCTCGGCAGTAAG | 1 620 1 654 |
表1 本研究所用引物信息
Table 1 Primers’ information used in this study
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product size/bp |
|---|---|---|
| 492-F 492-R 492-ZF 492-ZR | ATGAGATCTCTGTTTCTACGAG TCAACAGTCTCTCGGCAGTAAG GCTACGTAATGCATCATCATCATCATCATAGATCTCTGTTTCTACGA GCGGCCGCTCAACAGTCTCTCGGCAGTAAG | 1 620 1 654 |
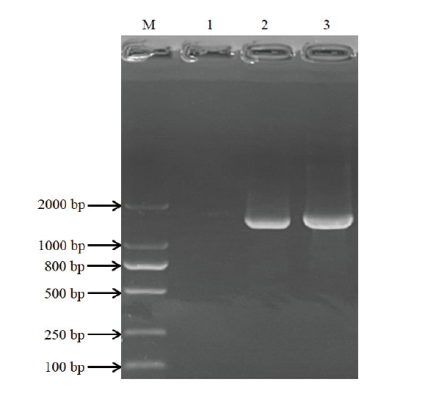
图1 少孢节丛孢菌Xj-2 AO-492基因的PCR扩增 M:DL2000 DNA marker;1:阴性对照;2-3:A0-492基因扩增产物
Fig. 1 PCR amplification of AO-492 gene of A. oligospora Xj-2 strain M: DL2000 DNA marker; 1: negative control; 2-3: amplified products of AO-492 gene
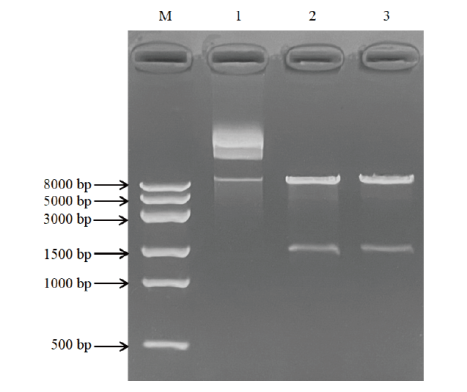
图4 重组质粒pPIC9K-AO492的双酶切鉴定 M:DL8000 DNA marker;1:未酶切的pPIC9K-AO492;2-3:pPIC9K-AO492双酶切产物
Fig. 4 Identification of recombinant plasmid pPIC9K-AO492 by double-nzyme digestion M: DL8000 DNA marker; 1: undigested pPIC9K-AO492; 2-3: double-enzyme digested products of pPIC9K-AO492
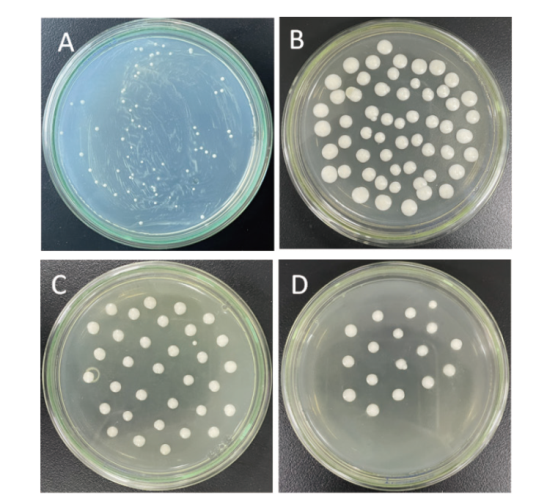
图5 重组酵母菌株的筛选 A:MD平板;B:含1 mg/mL G418的YPD培养基;C:含2 mg/mL G418的YPD培养基;D:含3 mg/mL G418的YPD培养基
Fig. 5 Screening of recombinant yeast strains A: MD plate; B: YPD medium containing 1 mg/mL G418; C: YPD medium containing 2 mg/mL G418; D: YPD medium containing 3 mg/mL G418

图6 重组蛋白ReAO-Z492的SDS-PAGE (A)及Westem blot(B)鉴定 M:蛋白分子质量标准;(A)1:pPIC9K空载体甲醇诱导72 h的培养液上清;2-6:甲醇分别诱导24、48、72、96和120 h的培养液上清;(B)1-2:纯化后浓缩的ReAO-Z492
Fig. 6 Identification of recombinant protein ReAO-Z492 by SDS-PAGE (A) and Western blot (B) M: Protein molecular quality standard ; 1: culture supernatant of pPIC9K empty vector induced by methanol for 72 h ; 2-6: culture supernatants induced by methanol for 24,48,72,96 and 120 h, respectively. (B) 1-2: Purified and concentrated ReAO-Z492
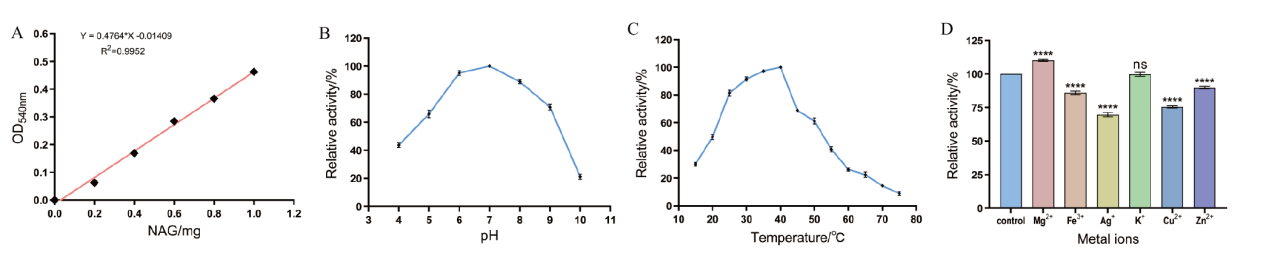
图7 重组几丁质酶AO-492相对酶活的测定 A:NAG标准曲线;B:不同pH对AO-492酶活性的影响;C:不同温度对AO-492酶活性的影响;D:不同金属离子对AO-492酶活性的影响。****表示P≤0.0001,ns表示P>0.05
Fig. 7 Determination of relative enzymatic activity of recombinant chitinase AO-492 A: NAG standard curve. B: The effect of different pH on the activity of AO-492. C: The effect of different temperatures on the activity of AO-492. D: The effect of different metal ions on the activity of AO-492. * * * * indicates P ≤ 0.0001, ns indicates P > 0.05

图8 ReAO-Z492对秀丽隐杆线虫的降解作用 对照组:A1-A3:用灭活的ReAO-Z492处理线虫0、3、6 h;试验组:B1-B3、C1-C3和D1-D3:ReAO-Z492处理线虫0、3、6 h。箭头所指为线虫部分体壁降解变化部位
Fig. 8 Degradation effect of ReAO-Z492 on C. elegans Control: A1-A3: C. elegans were treated with inactivated ReAO-Z492 for 0, 3, and 6 h. Experimentl: B1-B3, C1-C3 and D1-D3: C. elegans were treated with ReAO-Z492 for 0, 3, and 6 h. The arrow refers to the part of the body wall degradation of nematodes

图9 ReAO-Z492对秀丽隐杆线虫虫卵的降解作用 对照组:A1-A3:用灭活的ReAO-Z492处理虫卵0、6、12 h;试验组:B1-B3:ReAO-Z492处理虫卵0、6、12 h。箭头所指为虫卵卵壳降解部分
Fig. 9 Degradation effect of ReAO-Z492 on C. elegans eggs Control: A1-A3: C. elegans eggs were treated with inactivated ReAO-Z492 for 0, 6, and 12 h. Experiment: B1-B3: C. elegans eggs were treated with ReAO-Z492 for 0, 6, and 12 h. The arrow refers to the degradation part of egg shell
| [1] | Freitas LA, Savegnago RP, Menegatto LS, et al. Cluster analysis to explore additive-genetic patterns for the identification of sheep resistant, resilient and susceptible to gastrointestinal nematodes[J]. Vet Parasitol, 2022, 301: 109640. |
| [2] | Hou B, Yong R, Wuen JY, et al. Positivity rate investigation and anthelmintic resistance analysis of gastrointestinal nematodes in sheep and cattle in Ordos, China[J]. Animals, 2022, 12(7): 891. |
| [3] | Mendoza-de Gives P, Braga FR, de Araújo JV. Nematophagous fungi, an extraordinary tool for controlling ruminant parasitic nematodes and other biotechnological applications[J]. Biocontrol Sci Technol, 2022, 32(7): 777-793. |
| [4] | Zhang F, Boonmee S, Monkai J, et al. Drechslerella daliensis and D.xiaguanensis(Orbiliales, Orbiliaceae), two new nematode-trapping fungi from Yunnan, China[J]. Biodivers Data J, 2022, 10: e96642. |
| [5] | Niu XM, Zhang KQ. Arthrobotrys oligospora: a model organism for understanding the interaction between fungi and nematodes[J]. Mycology, 2011, 2(2): 59-78. |
| [6] |
Huang XW, Zhao NH, Zhang KQ. Extracellular enzymes serving as virulence factors in nematophagous fungi involved in infection of the host[J]. Res Microbiol, 2004, 155(10): 811-816.
pmid: 15567274 |
| [7] |
Yang JK, Tian BY, Liang LM, et al. Extracellular enzymes and the pathogenesis of nematophagous fungi[J]. Appl Microbiol Biotechnol, 2007, 75(1): 21-31.
pmid: 17318531 |
| [8] | Wernet N, Wernet V, Fischer R. The small-secreted cysteine-rich protein CyrA is a virulence factor participating in the attack of Caenorhabditis elegans by Duddingtonia flagrans[J]. PLoS Pathog, 2021, 17(11): e1010028. |
| [9] |
Gomaa EZ. Microbial chitinases: properties, enhancement and potential applications[J]. Protoplasma, 2021, 258(4): 695-710.
doi: 10.1007/s00709-021-01612-6 pmid: 33483852 |
| [10] | Qiu ST, Zhou SP, Tan Y, et al. Biodegradation and prospect of polysaccharide from crustaceans[J]. Mar Drugs, 2022, 20(5): 310. |
| [11] | Le B, Yang SH. Microbial chitinases: properties, current state and biotechnological applications[J]. World J Microbiol Biotechnol, 2019, 35(9): 144. |
| [12] | Yang JK, Yu Y, Li J, et al. Characterization and functional analyses of the chitinase-encoding genes in the nematode-trapping fungus Arthrobotrys oligospora[J]. Arch Microbiol, 2013, 195(7): 453-462. |
| [13] | Li C, Li XP, Bai CZ, et al. A chitinase with antifungal activity from naked oat(Avena chinensis)seeds[J]. J Food Biochem, 2019, 43(2): e12713. |
| [14] | Du JH, Duan S, Miao JY, et al. Purification and characterization of chitinase from Paenibacillus sp[J]. Biotechnol Appl Biochem, 2021, 68(1): 30-40. |
| [15] | Meerupati T, Andersson KM, Friman E, et al. Genomic mechanisms accounting for the adaptation to parasitism in nematode-trapping fungi[J]. PLoS Genet, 2013, 9(11): e1003909. |
| [16] | Yang JK, Wang L, Ji XL, et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation[J]. PLoS Pathog, 2011, 7(9): e1002179. |
| [17] | Chen W, Jiang X, Yang Q. Glycoside hydrolase family 18 chitinases: the known and the unknown[J]. Biotechnol Adv, 2020, 43: 107553. |
| [18] | Huang QS, Xie XL, Liang G, et al. The GH18 family of chitinases: their domain architectures, functions and evolutions[J]. Glycobiology, 2012, 22(1): 23-34. |
| [19] |
Houston DR, Recklies AD, Krupa JC, et al. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes[J]. J Biol Chem, 2003, 278(32): 30206-30212.
doi: 10.1074/jbc.M303371200 pmid: 12775711 |
| [20] |
Tjoelker LW, Gosting L, Frey S, et al. Structural and functional definition of the human chitinase chitin-binding domain[J]. J Biol Chem, 2000, 275(1): 514-520.
doi: 10.1074/jbc.275.1.514 pmid: 10617646 |
| [21] | Xie XH, Fu X, Yan XY, et al. A broad-specificity chitinase from Penicillium oxalicum k10 exhibits antifungal activity and biodegradation properties of chitin[J]. Mar Drugs, 2021, 19(7): 356. |
| [22] | Suryawanshi N, Eswari JS. Purification and characterization of chitinase produced by thermophilic fungi Thermomyces lanuginosus[J]. Prep Biochem Biotechnol, 2022, 52(9): 1087-1095. |
| [23] | Zargar V, Asghari M, Dashti A. A review on chitin and chitosan polymers: structure, chemistry, solubility, derivatives, and applications[J]. ChemBioEng Rev, 2015, 2(3): 204-226. |
| [24] | Moussian B. Chitin: structure, chemistry and biology[M]// Targeting Chitin-containing Organisms. Singapore: Springer, 2019: 5-18. |
| [25] | Tharanathan RN, Kittur FS. Chitin—the undisputed biomolecule of great potential[J]. Crit Rev Food Sci Nutr, 2003, 43(1): 61-87. |
| [26] | Chen Q, Peng DL. Nematode chitin and application[M]// Targeting Chitin-containing Organisms. Singapore: Springer, 2019: 209-219. |
| [27] |
Adrangi S, Faramarzi MA. From bacteria to human: a journey into the world of chitinases[J]. Biotechnol Adv, 2013, 31(8): 1786-1795.
doi: 10.1016/j.biotechadv.2013.09.012 pmid: 24095741 |
| [28] | Zhong WQ, Chen Y, Gong SS, et al. Enzymological properties and nematode-degrading activity of recombinant chitinase AO-379 of Arthrobotrys oligospora[J]. Kafkas Universitesi Veteriner Fakultesi Dergisi, 2019, 25(4): 435-444. |
| [29] |
贡莎莎, 孟庆玲, 乔军, 等. 少孢节丛孢菌XJ-A1几丁质酶AO-483基因的克隆及生物学活性[J]. 浙江农业学报, 2019, 31(2): 222-228.
doi: 10.3969/j.issn.1004-1524.2019.02.07 |
| Gong SS, Meng QL, Qiao J, et al. Gene cloning and bioactivity analysis of chitinase gene AO-483 from Arthrobotrys oligospora XJ-A1[J]. Acta Agric Zhejiangensis, 2019, 31(2): 222-228. | |
| [30] | Gong SS, Meng QL, Qiao J, et al. Biological characteristics of recombinant Arthrobotrys oligospora chitinase AO-801[J]. Korean J Parasitol, 2022, 60(5): 345-352. |
| [1] | 张瑶心, 王亮节, 郑文, 徐汉琴, 郑恋, 钟静. 产几丁质酶的无色杆菌ZWW8的发酵产酶及酶学性质研究[J]. 生物技术通报, 2021, 37(4): 96-106. |
| [2] | 郭继平, 马光, 王志杰, 齐善厚, 王宝梅, 苏长青. 一株解淀粉芽孢杆菌生防蛋白的鉴定及分析[J]. 生物技术通报, 2018, 34(1): 202-207. |
| [3] | 杨尉锦, 国果, 吴沁怡, 李妍, 付萍, 张勇. 家蝇几丁质酶基因MDCII重组表达质粒的构建及表达模式研究[J]. 生物技术通报, 2017, 33(2): 102-108. |
| [4] | 刘蒲临, 程德勇, 缪礼鸿. 产几丁质酶侧孢短芽孢杆菌的筛选及其酶学性质研究[J]. 生物技术通报, 2016, 32(6): 174-180. |
| [5] | 张圆, 周国旺, 李海涛, 刘荣梅, 赵一夔, 高继国. 苏云金芽胞杆菌chiA73基因的克隆、表达和酶活性分析[J]. 生物技术通报, 2015, 31(8): 147-152. |
| [6] | 李会琴;林炜铁;蔡小龙;李敬源;唐水水;. 对虾养殖水环境宏基因组Fosmid文库的构建[J]. , 2011, 0(06): 112-115. |
| [7] | 王广慧;. 含几丁质酶基因的双价防卫基因在植物抗逆基因工程中的应用[J]. , 2010, 0(10): 40-44. |
| [8] | 李世贵;顾金刚;姜瑞波;牛永春;. 生防木霉菌产几丁质酶特性研究[J]. , 2009, 0(04): 135-138. |
| [9] | 林建城;林俊兵;陈郁花;林秀春;. 南美白对虾体壁几丁质酶的理化特性研究[J]. , 2007, 0(04): 169-172. |
| [10] | 程茂高;乔卿梅;原国辉. 植物外源抗虫基因及其应用[J]. , 2004, 0(05): 18-20. |
| [11] | 刘欣洁;谭振波. 昆虫几丁质酶及其在植物抗虫品种改良中的应用[J]. , 2004, 0(03): 18-22. |
| [12] | 孟亮;李红双;金德敏;崔德才;王斌. 转几丁质酶基因黑杨的获得[J]. , 2004, 0(03): 48-51. |
| [13] | 欧阳石文;谢丙炎;赵开军;冯兰香. 转几丁质酶基因研究进展[J]. , 2001, 0(03): 28-31. |
| [14] | 李思经. 生物技术最新进展[J]. , 1997, 0(02): 25-26. |
| [15] | . 生物防治[J]. , 1996, 0(02): 82-85. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||