








生物技术通报 ›› 2025, Vol. 41 ›› Issue (1): 74-84.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0796
金素奎1( ), 国倩倩2, 刘巧泉1,2(
), 国倩倩2, 刘巧泉1,2( ), 高继平2(
), 高继平2( )
)
收稿日期:2024-08-17
出版日期:2025-01-26
发布日期:2025-01-22
通讯作者:
刘巧泉,男,博士,教授,研究方向:稻米品质遗传改良;E-mail: qqliu@yzu.edu.cn;作者简介:金素奎,男,博士,讲师,研究方向:稻米品质遗传改良;E-mail: skjin@yzu.edu.cn基金资助:
JIN Su-kui1( ), GUO Qian-qian2, LIU Qiao-quan1,2(
), GUO Qian-qian2, LIU Qiao-quan1,2( ), GAO Ji-ping2(
), GAO Ji-ping2( )
)
Received:2024-08-17
Published:2025-01-26
Online:2025-01-22
摘要:
【目的】为了应对日益增加的大规模遗传材料鉴定需求,急需建立一种快速、简易、高效、低成本的基因组DNA提取方法。【方法】根据水稻遗传鉴定的特点,对传统TPS法进行改进和简化。(1)将特制的钢珠加样条与全自动研磨仪结合使用,提高了研磨水稻叶片组织的速度;(2)使用TPS缓冲液室温研磨裂解,无需液氮冷冻研磨;(3)研磨后的组织匀浆仅需75℃水浴20 min即可完成裂解;(4)室温静置5 min后吸取粗提液上清5 μL至96孔PCR板内,使用排枪加入95 μL 灭菌的去离子水进行稀释,即完成基因组DNA的提取过程。【结果】简易TPS法提取基因组DNA的步骤减少至6步以内,省略了DNA沉淀、离心、洗涤、溶解等过程。提取过程简单、快速,可在30 min内完成96个样本的DNA提取。提取过程中使用的试剂种类较少且廉价易得、安全环保。通过优化DNA提取液的用量,用简易TPS法提取的基因组DNA开展的各类常规分子遗传鉴定的效果与CTAB法及传统TPS法相当。【结论】简易TPS法提高了水稻基因组DNA提取的效率,适合高通量的遗传鉴定。
金素奎, 国倩倩, 刘巧泉, 高继平. 一种水稻叶片基因组DNA简易提取方法[J]. 生物技术通报, 2025, 41(1): 74-84.
JIN Su-kui, GUO Qian-qian, LIU Qiao-quan, GAO Ji-ping. A Simplified Method for Extracting Genomic DNA from Rice Leaves[J]. Biotechnology Bulletin, 2025, 41(1): 74-84.

图1 简易TPS法提取水稻叶片基因组DNA的基本过程 A:使用2 mL离心管采集的水稻幼苗叶片;B:使用定制的钢珠加样条向离心管内加入研磨用的钢珠;C:使用全自动研磨仪振荡研磨叶片;D:裂解后的叶片组织匀浆
Fig. 1 Basic process of extracting genomic DNA from rice leaves using the simplified TPS method A: Rice seedling leaves were collected using a 2 mL centrifuge tube. B: Add grinding steel balls to the centrifugal tube using customized steel bar. C: Grinding the blade using an automatic grinding instrument. D: Homogenate of lysed leaf tissue
| 引物名称Primer name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') | 产物大小Product size/bp |
|---|---|---|---|
| OsACTIN1 | CCTTCAACACCCCTGCTATG | TGAGTAACCACGCTCCGTCA | 470 |
| STS-1 | CATGCTAGTAAGCAAAGGGCAACG | TTGCACGTCCAACTGTCCAAGC | 229 |
| HPT | GCTTTCAGCTTCGATGTAGGAGG | TTTCCACTATCGGCGAGTACTTC | 884 |
| OsNAP | AGTACCCACCCTCACAGCTC | AGTTGGTCTTGGTGCCCTTG | 319 |
| Wx pro | CTACTAGATCCGCTGCCGCC | GCAGGTCACAGCATTTATCTAAGC | 661 |
表1 本研究中所用引物的核苷酸序列
Table 1 Nucleotide sequences of primers used in this study
| 引物名称Primer name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') | 产物大小Product size/bp |
|---|---|---|---|
| OsACTIN1 | CCTTCAACACCCCTGCTATG | TGAGTAACCACGCTCCGTCA | 470 |
| STS-1 | CATGCTAGTAAGCAAAGGGCAACG | TTGCACGTCCAACTGTCCAAGC | 229 |
| HPT | GCTTTCAGCTTCGATGTAGGAGG | TTTCCACTATCGGCGAGTACTTC | 884 |
| OsNAP | AGTACCCACCCTCACAGCTC | AGTTGGTCTTGGTGCCCTTG | 319 |
| Wx pro | CTACTAGATCCGCTGCCGCC | GCAGGTCACAGCATTTATCTAAGC | 661 |
| 方法 Method | 使用试剂及仪器Reagents and instruments used | 步骤 Step | 耗时 Time/h | 纯度 Purity | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 液氮研磨 Liquid nitrogen grounding | CTAB | Tris | HCl | EDTA | NaCl | 氯仿 Chloroform | 异戊醇 Isoamylol | KCl | 异丙醇 Isopropanol | 乙醇 Ethanol | 离心机 Centrifuge | ||||
| CTAB法CTAB method | √ | √ | √ | √ | √ | √ | √ | √ | × | √ | √ | √ | ≥10 | 3-4 | 高 High |
| 传统TPS法Traditional TPS method | × | × | √ | √ | √ | × | × | × | √ | √ | √ | √ | ≥10 | 1-1.5 | 一般 Average |
| 简易TPS法Simplified TPS method | × | × | √ | √ | √ | × | × | × | √ | × | × | × | ≤6 | ≤0.5 | 低 Low |
表2 三种植物基因组DNA提取方法的比较
Table 2 Comparison of three methods for extracting genomic DNA from plants
| 方法 Method | 使用试剂及仪器Reagents and instruments used | 步骤 Step | 耗时 Time/h | 纯度 Purity | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 液氮研磨 Liquid nitrogen grounding | CTAB | Tris | HCl | EDTA | NaCl | 氯仿 Chloroform | 异戊醇 Isoamylol | KCl | 异丙醇 Isopropanol | 乙醇 Ethanol | 离心机 Centrifuge | ||||
| CTAB法CTAB method | √ | √ | √ | √ | √ | √ | √ | √ | × | √ | √ | √ | ≥10 | 3-4 | 高 High |
| 传统TPS法Traditional TPS method | × | × | √ | √ | √ | × | × | × | √ | √ | √ | √ | ≥10 | 1-1.5 | 一般 Average |
| 简易TPS法Simplified TPS method | × | × | √ | √ | √ | × | × | × | √ | × | × | × | ≤6 | ≤0.5 | 低 Low |
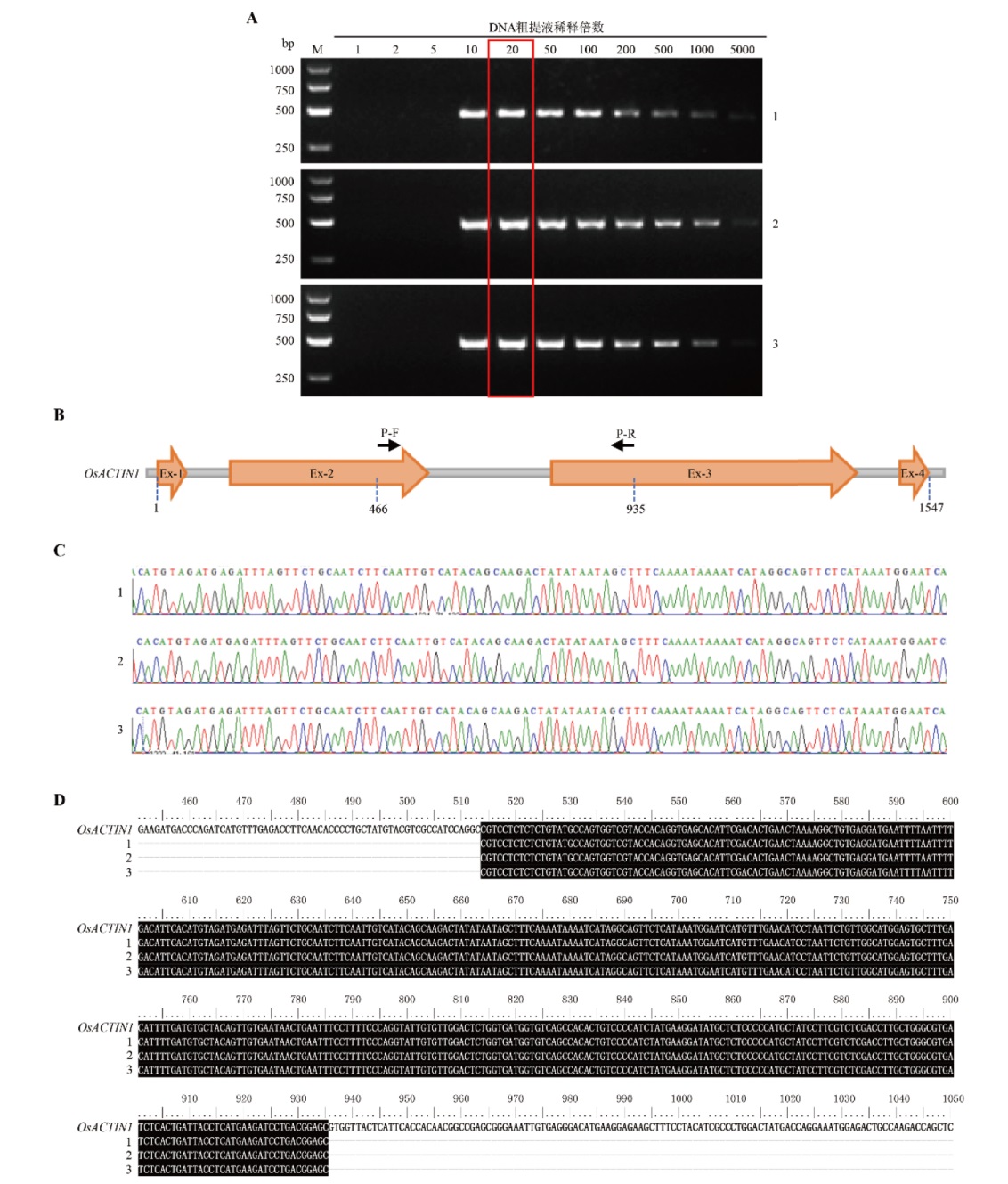
图2 简易TPS法提取水稻叶片基因组DNA的PCR条件探索与优化 A:以日本晴幼苗的叶片为材料提取基因组DNA,以水稻OsACTIN1基因片段作为PCR扩增目的基因,PCR反应体系及条件如材料方法中所述; M为DNA marker(GoldBand DL5000(翊圣生物));顶端数字表示DNA粗提液稀释倍数;图右侧数字表示不同的单株重复;B:水稻OsACTIN1基因序列的结构示意图。其中橙色带方向框以及字母Ex-1/2/3/4表示外显子,黑色箭头以及字母P-F/R分别表示PCR扩增中使用的正向和反向引物,虚竖线及其下方的数字表示碱基位置;C:OsACTIN1的PCR产物进行测序所获得部分峰图。对基因组DNA稀释20倍条件下获得的PCR产物进行测序,测序引物为OsACTIN1基因PCR扩增的正向引物;D:对测序获得的序列与OsACTIN1的基因序列进行比对。黑色背景的字母表示相同的碱基
Fig. 2 Exploration and optimization of PCR conditions for extracting genomic DNA from rice leaves by the simplified TPS method A: The genomic DNA was extracted from the leaves of Nipponbare seedlings, and the rice OsACTIN1 gene fragment was used as the target gene for PCR amplification. The PCR reaction system and conditions were described in the Material and Method section. M stands for DNA marker(GoldBand DL5000(YEASEN BioTech)); the numbers at the top of the figure refers to the dilution ratio of DNA crude extract; the numbers on the left of the figure refer to band size(bp)of DNA marker; the numbers on the right of the figure refer to different individual replicates. B: Structural diagram of rice OsACTIN1 gene. The orange boxes with direction and the letters Ex-1/2/3/4 indicate exons, the black arrows with the letters P-F/R above indicate forward and reverse primers used in PCR amplification, respectively, and the dashed vertical lines with the number below indicate base positions. C: The partial peak maps of sequenced PCR products. The PCR products were obtained and sequenced under the 20-fold dilution of the genomic DNA, and the sequencing primer was the forward primer of OsACTIN1 gene used in PCR amplification. D: The sequences obtained by sequencing were aligned with the OsACTIN1gene. The letters on the black background indicate the same bases
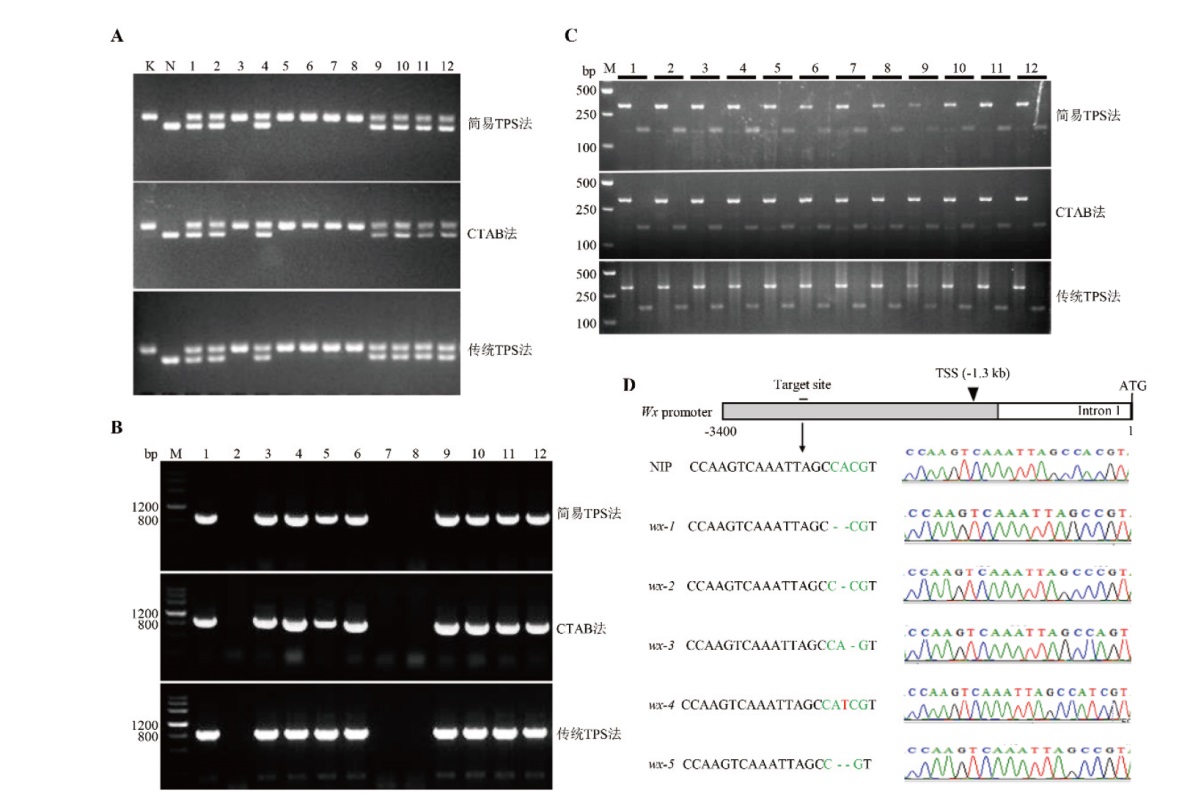
图3 简易TPS法与CTAB法、传统TPS法提取的水稻基因组DNA用于遗传鉴定的比较 A:用Koshihikari(K)、Nona Bokra(N)及其F2个体提取基因组DNA,并用分子标记STS-1检测基因型。K和N分别表示2个亲本,数字1-12分别表示12个不同的单株;B:以日本晴背景的OsNAC24 gD-OE过表达转基因植株为材料,检测HPT基因。M表示DNA marker,本实验使用的DNA marker为DNA marker III(天根生化),数字1-12表示12个不同的单株,其中第2、7、8号3个单株为阴性单株;C:以野生型日本晴为材料,扩增OsNAP基因片段,并使用BamH I酶切PCR产物。M表示DNA marker,泳道顶端数字1-12表示12个不同的单株,每个单株左侧的泳道为未酶切的PCR产物,右侧的泳道为被BamHI酶切后的PCR产物;D:对日本晴中编辑Wx基因启动子的转基因植株进行的测序鉴定。图的上部是Wx基因的启动子结构示意图,图的下方左侧为测序后的序列,图的下右侧为对应的测序峰图。TSS表示转录起始位点,ATG表示起始密码子。NIP:日本晴
Fig. 3 Comparison of rice genomic DNA extracted by the simplified TPS method, CTAB method, and traditional TPS method in genetic identification A: The genomic DNA was extracted from Koshihikari(K), Nona Bokra(N)and their F2 individuals, and the molecular marker STS-1 was used in genotyping. K and N refer to two parents respectively, and the number 1 to 12 refer to 12 individual plants. B: The HPT gene was detected in the OsNAC24 gD-OE transgenic plants under Nipponbare background. M refers to DNA marker, the DNA marker used in this experiment was DNA marker III(TIANGEN Biochemistry), and the number 1 to 12 refer to 12 individual plants, among which, 3 plants No. 2, 7, and 8 were negative. C: The wild-type Nipponbare plants were used to amplify the OsNAP gene, then the PCR product was digested by BamH I. M refers to DNA marker, the DNA marker used in this experiment was GoldBand DL5000(YEASEN BioTech); and the number from 1 to 12 at the top of the lane refer to 12 individual plants. The lane on the left of each single plant is the PCR product without digestion, and the lane on the right is the PCR product digested by the BamHI. D: Identification of transgenic plants with edited Wx promoter by sequencing in Nipponbare. The upper part of the figure is a schematic diagram of the Wx promoter, the lower left part of the figure are the sequences of target, and the lower right part of the figure is the corresponding sequencing peak. TSS stands for the transcription start site and ATG refers to the start codon. NIP: Nipponbare
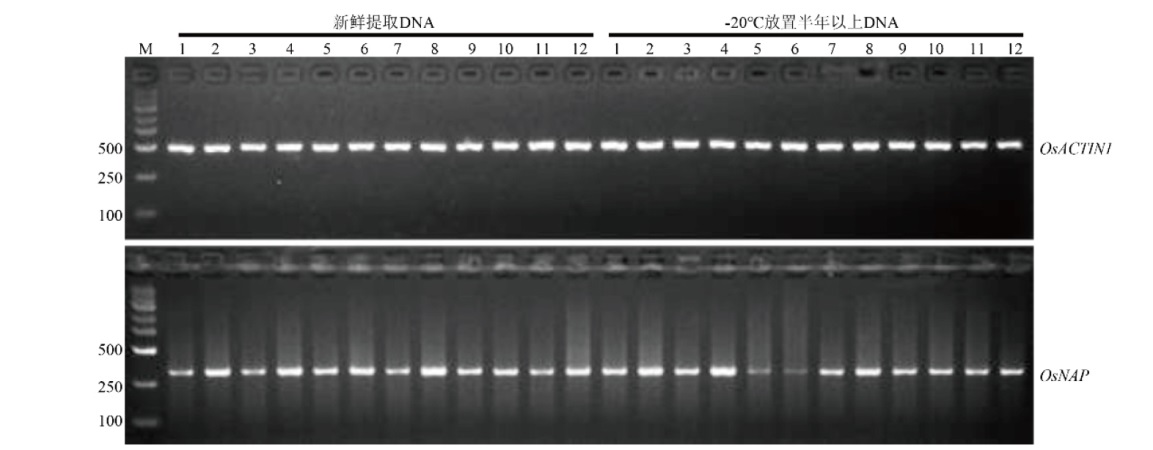
图4 简易TPS法提取的基因组DNA的稳定性检测 分别用新鲜提取的和在-20℃保存半年以上的日本晴基因组DNA检测OsACTIN1和OsNAP基因。M表示DNA marker,本实验使用的DNA marker为GoldBand DL5000(翊圣生物),数字1-12表示12个不同的单株
Fig. 4 Stability detection of genomic DNA extracted by simplified TPS method The detection of OsACTIN1 and OsNAP genes was performed using genomic DNA of Nipponbare freshly extracted and stored at -20℃ for more than six months, respectively. M refers to DNA marker, the DNA marker used in this experiment is GoldBand DL5000(YEASEN BioTech), and the number 1 to 12 refer to 12 different individual plants
| [1] | Zheng KX, Cai YC, Chen WJ, et al. Development, identification, and application of a germplasm specific SCAR marker for Dendrobium officinale kimura et migo[J]. Front Plant Sci, 2021, 12: 669458. |
| [2] | 张紫量, 李旭鹏, 申君毅, 等. 植物DNA提取方法的研究进展[J]. 农业与技术, 2024, 44(14): 166-170. |
| Zhang ZL, Li XP, Shen JY, et al. Research progress of plant DNA extraction methods[J]. Agric Technol, 2024, 44(14): 166-170. | |
| [3] | Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton(Gossypium spp.)genomic DNA suitable for RFLP or PCR analysis[J]. Plant Mol Biol Report, 1993, 11(2): 122-127. |
| [4] | 李岩, 于海琳, 马晓彤, 等. 对CTAB法提取菠菜基因组DNA实验的优化设计[J]. 化工管理, 2023(14): 15-17. |
| Li Y, Yu HL, Ma XT, et al. Conditions research on CTAB extraction of spinach genome DNA[J]. Chem Enterp Manag, 2023(14): 15-17. | |
| [5] |
李金璐, 王硕, 于婧, 等. 一种改良的植物DNA提取方法[J]. 植物学报, 2013, 48(1): 72-78.
doi: 10.3724/SP.J.1259.2013.00072 |
| Li JL, Wang S, Yu J, et al. A modified CTAB protocol for plant DNA extraction[J]. Chin Bull Bot, 2013, 48(1): 72-78. | |
| [6] | 王珅, 范保杰, 刘长友, 等. 绿豆愈伤组织基因组DNA最佳提取方法的分析[J/OL]. 分子植物育种, 2024, 1-13. |
| Wang K, Fan BJ, Liu CY, et al. Analysis of the best extraction methods of genomic DNA from mung bean callus[J/OL]. Molecular Plant Breeding, 2024, 1-13. | |
| [7] | 张凤娟, 张满良, 朱水芳. 一种改进的水稻总DNA的快速提取方法[J]. 植物检疫, 2004, 18(6): 330-332. |
| Zhang FJ, Zhang ML, Zhu SF. An improved rapid method of plant total DNA extraction[J]. Plant Quar, 2004, 18(6): 330-332. | |
| [8] | 毕继安, 王芳, 张国芳, 等. 药用野生稻基因组DNA提取方法比较[J]. 安徽农业科学, 2023, 51(14): 86-89. |
| Bi JA, Wang F, Zhang GF, et al. Comparison of different extraction methods of genomic DNA from Oryza officinalis[J]. J Anhui Agric Sci, 2023, 51(14): 86-89. | |
| [9] | 苏代群. 水稻SSR分析快速提取总DNA法[J]. 种子世界, 2013(11): 28-30. |
| Su DQ. Rapid extraction of total DNA for rice SSR analysis[J]. Seed World, 2013(11): 28-30. | |
| [10] | 田孟祥, 宫彦龙, 张时龙, 等. 简便烘干在水稻叶片DNA大量提取中的应用[J]. 河南农业科学, 2020, 49(2): 52-57. |
| Tian MX, Gong YL, Zhang SL, et al. Application of simple drying in extraction of large amount of DNA from rice leaves[J]. J Henan Agric Sci, 2020, 49(2): 52-57. | |
| [11] | 马文东. 水稻基因组DNA提取方法的研究[J]. 黑龙江农业科学, 2014(1): 7-11. |
| Ma WD. Study on extraction method of rice genomic DNA[J]. Heilongjiang Agric Sci, 2014(1): 7-11. | |
| [12] | 蒋小刚, 王华, 周武先, 等. 白术叶片DNA提取方法的优化[J]. 安徽农业科学, 2023, 51(11): 128-131. |
| Jiang XG, Wang H, Zhou WX, et al. Optimization of DNA extraction method for Atractylodes macrocephala leaves[J]. J Anhui Agric Sci, 2023, 51(11): 128-131. | |
| [13] | 杨祥波. 优化的SDS法提取刺五加基因组的研究[J]. 吉林农业, 2015(21): 72-73. |
| Yang XB. Study on the extraction of Acanthopanax acanthopanax genome by optimized SDS method[J]. Agric Jilin, 2015(21): 72-73. | |
| [14] | 侯泽菁, 常廼滔. 用改良SDS法提取适于PCR扩增的小麦基因组DNA[J]. 江苏农业科学, 2014, 42(5): 49-51. |
| Hou ZJ, Chang NT. Using modified SDS method to extract wheat genomic DNA suitable for PCR amplification[J]. Jiangsu Agric Sci, 2014, 42(5): 49-51. | |
| [15] | 廖云蓉. 使用改良的SDS法提取银杏不同器官DNA的研究[J]. 现代园艺, 2012(11): 5, 78. |
| Liao YR. Study on the DNA extraction from different organs of Ginkgo biloba by improved SDS method[J]. Xiandai Hortic, 2012(11): 5, 78. | |
| [16] | 穆春华, 张发军, 李文才, 等. 玉米叶片基因组快速提取方法研究[J]. 玉米科学, 2010, 18(3): 170-172. |
| Mu CH, Zhang FJ, Li WC, et al. A method of genomic DNA extraction of maize[J]. J Maize Sci, 2010, 18(3): 170-172. | |
| [17] |
张友昌, 冯常辉, 别墅, 等. 改进的TPS法: 一种棉花叶片DNA快速提取方法[J]. 棉花学报, 2016, 28(4): 413-417.
doi: 10.11963/issn.1002-7807.201604014 |
|
Zhang YC, Feng CH, Bie S, et al. An improved TPS method for rapid DNA extraction from cotton leaves[J]. Cotton Sci, 2016, 28(4): 413-417.
doi: 10.11963/issn.1002-7807.201604014 |
|
| [18] | 王蕾, 赵新涛, 许梦琦, 等. 花生叶片DNA快速提取方法的优化[J]. 山东农业科学, 2014, 46(7): 11-14. |
| Wang L, Zhao XT, Xu MQ, et al. Optimization of rapid DNA extraction methods from peanut leaves[J]. Shandong Agric Sci, 2014, 46(7): 11-14. | |
| [19] |
姚丹, 闫伟, 关淑艳, 等. 高盐低pH值法提取大豆不同组织DNA的效果[J]. 河南农业科学, 2009, 38(12): 50-54.
doi: 10.3969/j.issn.1004-3268.2009.12.015 |
| Yao D, Yan W, Guan SY, et al. Extraction effect of genomic DNA from different tissues of soybean with high-salt low-pH methods[J]. J Henan Agric Sci, 2009, 38(12): 50-54. | |
| [20] | 许理文, 王凤格, 赵久然, 等. 高盐低pH值法提取玉米基因组DNA的研究[J]. 玉米科学, 2009, 17(1): 59-61, 70. |
| Xu LW, Wang FG, Zhao JR, et al. Studies on genomic DNA extraction of maize with high salt, low pH method[J]. J Maize Sci, 2009, 17(1): 59-61, 70. | |
| [21] | 郭娴, 张寒霜, 赵俊丽, 等. 高盐低pH值法提取棉花基因组DNA研究[C]// 中国棉花学会. 中国棉花学会2011年年会论文汇编, 2011: 1. |
| Guo X, Zhang H, Zhao J, et al. Studies on Genomic DNA Extraction of Cotton with High Salt, Low pH Method[C]. China Cotton Society, China Cotton Society 2011 annual meeting paper compilation, 2011: 1. | |
| [22] | 杨学珍, 刘晓雪, 宋健, 等. 改良高盐高pH法提取大豆DNA[J]. 大豆科学, 2020, 39(4): 549-554. |
| Yang XZ, Liu XX, Song J, et al. Extraction of soybean DNA by improved high salt and high pH method[J]. Soybean Sci, 2020, 39(4): 549-554. | |
| [23] | 孙利萍, 贾芝琪, 胡建斌, 等. 碱裂解法快速提取番茄DNA的研究[J]. 河南农业大学学报, 2012, 46(2): 136-138. |
| Sun LP, Jia ZQ, Hu JB, et al. Study on the rapid alkaline Lysis method of extracting tomato DNA[J]. J Henan Agric Univ, 2012, 46(2): 136-138. | |
| [24] | 吴则东, 王华忠, 倪洪涛. 不同碱裂解法快速提取甜菜大群体DNA的研究[J]. 中国糖料, 2013, 35(3): 32-34. |
| Wu ZD, Wang HZ, Ni HT. A rapid method for extract beet large group DNA in different alkaline Lysis[J]. Sugar Crops China, 2013, 35(3): 32-34. | |
| [25] |
王涛, 王超楠, 张红, 等. 大白菜基因组DNA快速提取方法的研究[J]. 华北农学报, 2017, 32(6): 67-72.
doi: 10.7668/hbnxb.2017.06.010 |
| Wang T, Wang CN, Zhang H, et al. Study on rapid extraction of genomic DNA from Chinese cabbage[J]. Acta Agric Boreali Sin, 2017, 32(6): 67-72. | |
| [26] | 赵国珍, 贾育林, 严宗卜, 等. 一种高效便捷的水稻DNA提取法及其应用[J]. 中国水稻科学, 2012, 26(4): 495-499. |
|
Zhao GZ, Jia YL, Yan ZB, et al. An efficient, economic, and rapid rice DNA extraction method and its application[J]. Chin J Rice Sci, 2012, 26(4): 495-499.
doi: 10.3969/j.issn.1001-7216.2012.04.016 |
|
| [27] | 李柯, 赵昆昆, 宁龙龙, 等. 花生DNA快速提取方法及应用[J]. 山东农业科学, 2019, 51(9): 68-72. |
| Li K, Zhao KK, Ning LL, et al. A rapid extraction method of peanut DNA and its application[J]. Shandong Agric Sci, 2019, 51(9): 68-72. | |
| [28] | 曹文波, 郑璐璐, 谢文海. 一种提取植物基因组DNA的方法——改良尿素法[J]. 华中师范大学学报: 自然科学版, 2008, 42(3): 448-451. |
| Cao WB, Zheng LL, Xie WH. The modification urea method: an improved method for plant DNA isolation[J]. J Huazhong Norm Univ Nat Sci, 2008, 42(3): 448-451. | |
| [29] | 郑卓, 李健, 圣忠华, 等. 高粱总DNA不同提取方法的比较[J]. 安徽农业科学, 2007, 35(36): 11758-11759. |
| Zheng Z, Li J, Sheng ZH, et al. Comparison of different extraction methods of total DNA from sorghum[J]. J Anhui Agric Sci, 2007, 35(36): 11758-11759. | |
| [30] | 石庆华, 姚正培, 张桦, 等. 4种提取鹰嘴豆基因组DNA方法的比较[J]. 新疆农业大学学报, 2009, 32(1): 64-67. |
| Shi QH, Yao ZP, Zhang H, et al. Comparison of four methods of DNA extraction from chickpea[J]. J Xinjiang Agric Univ, 2009, 32(1): 64-67. | |
| [31] | Selz J, Adam NR, Magrini CEM, et al. A field-capable rapid plant DNA extraction protocol using microneedle patches for botanical surveying and monitoring[J]. Appl Plant Sci, 2023, 11(3): e11529. |
| [32] | 王润语. 植物DNA提取试剂盒的研究及在检测转基因稻米上的应用[J]. 生物技术世界, 2016, 13(1): 1-3. |
| Wang RY. Study on plant extraction kit and its application in detecting transgenic rice[J]. Biotech World, 2016, 13(1): 1-3. | |
| [33] |
Güssow D, Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction[J]. Nucleic Acids Res, 1989, 17(10): 4000.
doi: 10.1093/nar/17.10.4000 pmid: 2734114 |
| [34] | Jin SK, Xu LN, Leng YJ, et al. The OsNAC24-OsNAP protein complex activates OsGBSSI and OsSBEI expression to fine-tune starch biosynthesis in rice endosperm[J]. Plant Biotechnol J, 2023, 21(11): 2224-2240. |
| [35] | 张红, 王超楠, 范伟强, 等. 植物基因组DNA的高效提取方法[J/OL]. 分子植物育种, 2022, 1-14. |
| Zhang H, Wang CN, Fan WQ, et al. Efficient extraction of plant genomic DNA[J/OL]. Molecular Plant Breeding, 2022, 1-14. | |
| [36] | 肖帅, 李鳞霞, 李海鸥. 一种高效的植物DNA提取和PCR扩增体系建立[J]. 亚热带植物科学, 2018, 47(3): 207-210. |
| Xiao S, Li LX, Li HO. An efficient plant DNA extraction and PCR amplification system[J]. Subtrop Plant Sci, 2018, 47(3): 207-210. | |
| [37] | 朱先飞, 韩仁长, 黄冠, 等. 油菜DNA快速高通量提取方法改良及应用[J]. 园艺与种苗, 2023, 43(8): 83-85. |
| Zhu XF, Han RH, Huang G, et al. Improvement and application of rapid and high-throughput DNA extraction method for rapeseed[J]. Hortic Seed, 2023, 43(8): 83-85. | |
| [38] | 田孟祥, 张时龙, 余本勋, 等. 一种应用PCR缓冲液快速制备水稻DNA模板的方法[J]. 分子植物育种, 2015, 13(2): 438-442. |
| Tian MX, Zhang SL, Yu BX, et al. A method by using PCR buffer to rapid prepare rice DNA template[J]. Mol Plant Breed, 2015, 13(2): 438-442. | |
| [39] | 王攀. 植物分子标记高通量快速检测技术的研究进展[J]. 中国种业, 2024(7): 17-22. |
| Wang P. Research progress on high-throughput rapid detection technology for plant molecular markers[J]. China Seed Ind, 2024(7): 17-22. | |
| [40] | 陈晓军, 王敬东, 宋海丽, 等. 一种简单、极快的植物叶片DNA提取方法[J]. 种子, 2018, 37(11): 26-29, 34. |
| Chen XJ, Wang JD, Song HL, et al. A simple and rapid method of DNA extraction from plant leaf[J]. Seed, 2018, 37(11): 26-29, 34. |
| [1] | 杨秀清, 陈彦梅, 吴瑞薇, 王保玉, 韩作颖. 煤地质环境微生物总基因组DNA提取方法的优化[J]. 生物技术通报, 2018, 34(9): 177-183. |
| [2] | 杜文凯, 袁素霞, 胡凤荣. 一种快速检测岷江百合总RNA样本中基因组DNA残留的方法[J]. 生物技术通报, 2018, 34(10): 81-86. |
| [3] | 李海洋, 王飞, 雷红涛, 张璇, 陈珂. 硅羟基磁珠的制备及全基因组DNA提取优化[J]. 生物技术通报, 2017, 33(6): 223-229. |
| [4] | 黄新凤, 叶金波, 刘建军. 质谱技术在DNA甲基化研究中的应用[J]. 生物技术通报, 2015, 31(11): 112-120. |
| [5] | 迟婧,耿丽丽,高继国,束长龙,张杰,. 植物叶片基因组DNA快速提取方法[J]. 生物技术通报, 2014, 30(9): 51-57. |
| [6] | 张彦蕊;束长龙;宋福平;黄大昉;张杰;. 一种简单、快速的苏云金芽胞杆菌基因组DNA提取方法[J]. , 2012, 0(11): 197-201. |
| [7] | 王克平;叶江;张惠展;. 迟钝爱德华氏菌RNA提取及痕量基因组DNA去除方法探究[J]. , 2012, 0(06): 179-182. |
| [8] | 范慧慧;李明云;. 香鱼基因组DNA提取方法的优化[J]. , 2012, 0(05): 158-162. |
| [9] | 李莉;李春梅;谢旭阳;. 三种提取柱状黄杆菌基因组DNA方法的比较[J]. , 2012, 0(05): 163-166. |
| [10] | 陈丽丽;张鹏;黄新;朱水芳;. 玉米基因组DNA提取及浓度测定方法评价[J]. , 2011, 0(12): 70-74. |
| [11] | 徐飞;成述儒;罗玉柱;. FTA卡和普通定性滤纸提取DNA方法研究[J]. , 2011, 0(11): 221-224. |
| [12] | 蔡翠霞;肖维威;康文杰;周琳华;吴永彬;郑远金;马文丽;. 改良Chelex-100法快速提取转基因农产品DNA[J]. , 2011, 0(10): 210-214. |
| [13] | 何建文;韩建林;罗玉柱;. 利用不同方法从深加工牦牛肉产品中提取基因组DNA效果的比较[J]. , 2010, 0(10): 162-167. |
| [14] | 谢放;陈京津;朱子雄;. 三种提取冬虫夏草菌丝体基因组DNA方法的比较[J]. , 2010, 0(08): 134-136. |
| [15] | 李敏俐;关善辉;陆祖宏;. 染色质免疫沉淀试验中基因组DNA超声破碎条件优化策略[J]. , 2010, 0(05): 121-125. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||