








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 184-197.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0504
收稿日期:2025-05-16
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
伍国强,男,博士,教授,博士生导师,研究方向 :植物逆境生理与基因工程;E-mail: gqwu@lut.edu.cn作者简介:任云儿,女,硕士研究生,研究方向 :生物工程;E-mail: 954466727@qq.com
基金资助:
REN Yun-er( ), WU Guo-qiang(
), WU Guo-qiang( ), CHENG Bin, WEI Ming
), CHENG Bin, WEI Ming
Received:2025-05-16
Published:2026-01-26
Online:2026-02-04
摘要:
目的 自噬是维持生物体内细胞稳态的重要降解途径,在调控植物响应逆境胁迫中起着重要作用。挖掘和鉴定甜菜(Beta vulgaris L.)自噬相关基因(autophagy-related genes, ATGs)家族成员,并分析其在盐胁迫下的表达模式,为探究BvATGs在逆境下的功能提供理论依据。 方法 利用生物信息学方法从甜菜基因组中鉴定BvATGs基因家族成员,分析其蛋白质理化性质、染色体分布、系统发育、基因结构、保守基序、顺式调控元件和共线性;采用RT-qPCR分析BvATGs在盐胁迫下的表达模式;采用PEG介导法瞬时转化拟南芥原生质体,以确定BvATG4和BvATG6a-1亚细胞定位;构建BvATG4和BvATG6a-1过表达载体并遗传转化拟南芥。 结果 鉴定出51个BvATGs基因,分为20个亚家族;其中,48个BvATGs不均地分布在9条染色体上,而3个基因(BvATG1a、BvATG1d和BvATG1k)未定位在染色体;同一亚家族内的BvATGs基因具有相似的基因结构和保守域。BvATGs编码蛋白质氨基酸数为84‒2 467 aa,分子质量为10.21‒277.30 kD,大部分为亲水性蛋白质;88.2%的BvATGs成员定位于细胞质、细胞核和叶绿体。通过甜菜物种内共线性分析发现BvATGs有3对同源基因,物种间共线性显示BvATGs在水稻和拟南芥中分别存在9对和30对同源基因;在BvATGs启动子区含有大量的光响应元件、激素响应元件和逆境胁迫响应元件。RT-qPCR分析显示,盐胁迫处理下,12个BvATG基因在叶和根中的表达量均有不同程度上调。BvATG4主要定位于细胞核和细胞质,BvATG6a-1主要定位于细胞质及内质网。与野生型拟南芥相比,转基因株系中BvATG4和BvATG6a-1基因相对表达水平均显著增加。 结论 从甜菜全基因组中鉴定出51个BvATGs基因家族成员,其中12个基因不同程度地受盐胁迫诱导和上调。BvATG4定位在细胞核和细胞质,而BvATG6a-1定位在细胞质和内质网。进一步将BvATG4和BvATG6a-1转入拟南芥,分别获得高表达的转基因株系OE4和OE2。为甜菜耐盐基因资源挖掘与利用奠定基础。
任云儿, 伍国强, 成斌, 魏明. 甜菜BvATGs基因家族全基因组鉴定及盐胁迫下表达模式分析[J]. 生物技术通报, 2026, 42(1): 184-197.
REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress[J]. Biotechnology Bulletin, 2026, 42(1): 184-197.
| 基因名称 Gene name | 上游引物 Forward primer sequence (5′‒3′) | 下游引物 Reverse primer sequence (5′‒3′) |
|---|---|---|
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
| BvATG1e | CAATCTGTCTGGCTTATTCGATG | CGCTCGAAGTTTAGCAACCC |
| BvATG1h | TTTGCCGGAACACCTTAGACA | CCTTAGCACCATCATATCCCT |
| BvATG3 | CCATGCTCGCAAAACGGTA | GTCAACTTCTGGCTCAACCC |
| BvATG4 | TCGTCGTCAACTAGGGCTTC | CTCCTCCGAACGATGCTCA |
| BvATG6a-1 | GTCTCCCTAAAGTTCCGGTTG | TTTACAGGCCCAAACAGGT |
| BvATG8b | CCTACTGGAGCAATCATGTCT | CTCCACAGCAAGATACCCGAA |
| BvATG8c | GCTCCAACATCCTCTCGAAC | TGCCCCACAGTTAAATCAGC |
| BvATG8d | TCCTGAGAAAGCCATATTCGT | TAGCAATGCACCTCAATCCAA |
| BvATG8e | TGCCAAGTATCCTGATCGAGT | ATATAAATAGAGCTTTGCCCGGAG |
| BvATG10 | CCTCTCTTCACTTCCCACGA | TTGCAGGATCAATACGCTCT |
| BvATG16-2 | CAGTGCAGCTTATGATCGAAC | CAAAGCCGGAGATTCCCAT |
| BvATG20c | TCCCGAGATTTCCACTACTGC | AAGGATCGAAACATAGCGTCA |
表1 本研究所用RT-qPCR引物序列
Table 1 Primer sequences for RT-qPCR in this study
| 基因名称 Gene name | 上游引物 Forward primer sequence (5′‒3′) | 下游引物 Reverse primer sequence (5′‒3′) |
|---|---|---|
| BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
| BvATG1e | CAATCTGTCTGGCTTATTCGATG | CGCTCGAAGTTTAGCAACCC |
| BvATG1h | TTTGCCGGAACACCTTAGACA | CCTTAGCACCATCATATCCCT |
| BvATG3 | CCATGCTCGCAAAACGGTA | GTCAACTTCTGGCTCAACCC |
| BvATG4 | TCGTCGTCAACTAGGGCTTC | CTCCTCCGAACGATGCTCA |
| BvATG6a-1 | GTCTCCCTAAAGTTCCGGTTG | TTTACAGGCCCAAACAGGT |
| BvATG8b | CCTACTGGAGCAATCATGTCT | CTCCACAGCAAGATACCCGAA |
| BvATG8c | GCTCCAACATCCTCTCGAAC | TGCCCCACAGTTAAATCAGC |
| BvATG8d | TCCTGAGAAAGCCATATTCGT | TAGCAATGCACCTCAATCCAA |
| BvATG8e | TGCCAAGTATCCTGATCGAGT | ATATAAATAGAGCTTTGCCCGGAG |
| BvATG10 | CCTCTCTTCACTTCCCACGA | TTGCAGGATCAATACGCTCT |
| BvATG16-2 | CAGTGCAGCTTATGATCGAAC | CAAAGCCGGAGATTCCCAT |
| BvATG20c | TCCCGAGATTTCCACTACTGC | AAGGATCGAAACATAGCGTCA |
| 引物用途 Primer purpose | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene clone | BvATG4-F-Xho I | GAGGACACGCTCGAGATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Sma I | CTTTGTAATCCCCGGGGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Xho I | TTTGGAGAGGACACGCTCGAGATGAAGGAGAAGCAGCTTG | |
| BvATG6a-1-R-Sma I | TCATCTTTGTAATCCCCGGGTTGACCAGATGGAAATCGAG | |
转基因拟南芥PCR检测 Transgenic A. thaliana PCR detection | 35S-F | GACGCACAATCCCACTATCC |
| BvATG4-R | GAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-R | TTGACCAGATGGAAATCGAG | |
内参基因 Refenrence gene | AtUBQ10-F | AGATCCAGGACAAGGAAGGTATTC |
| AtUBQ10-R | CGCAGGACCAAGTGAAGAGTAG | |
亚细胞定位 Subcellular localization | BvATG4-F-Hind III | GGACAGCCCAAGCTTATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Nco I | TTGCTCACCATGGATCCGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Hind III | CATGGAGGCCAGTGAATTCATGAAGGAGAAGCAGCTTGATG | |
| BvATG6a-1-R-Nco I | TCGAGCTCGATGGATCCTTGACCAGATGGAAATCGAGAATC |
表2 本研究中所用的引物序列
Table 2 Primers used in the study
| 引物用途 Primer purpose | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
基因克隆 Gene clone | BvATG4-F-Xho I | GAGGACACGCTCGAGATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Sma I | CTTTGTAATCCCCGGGGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Xho I | TTTGGAGAGGACACGCTCGAGATGAAGGAGAAGCAGCTTG | |
| BvATG6a-1-R-Sma I | TCATCTTTGTAATCCCCGGGTTGACCAGATGGAAATCGAG | |
转基因拟南芥PCR检测 Transgenic A. thaliana PCR detection | 35S-F | GACGCACAATCCCACTATCC |
| BvATG4-R | GAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-R | TTGACCAGATGGAAATCGAG | |
内参基因 Refenrence gene | AtUBQ10-F | AGATCCAGGACAAGGAAGGTATTC |
| AtUBQ10-R | CGCAGGACCAAGTGAAGAGTAG | |
亚细胞定位 Subcellular localization | BvATG4-F-Hind III | GGACAGCCCAAGCTTATGAAGAACTTGTTTGAGAGTGCTGG |
| BvATG4-R-Nco I | TTGCTCACCATGGATCCGAGGAGTTGCCATTCGTCCTCTTG | |
| BvATG6a-1-F-Hind III | CATGGAGGCCAGTGAATTCATGAAGGAGAAGCAGCTTGATG | |
| BvATG6a-1-R-Nco I | TCGAGCTCGATGGATCCTTGACCAGATGGAAATCGAGAATC |
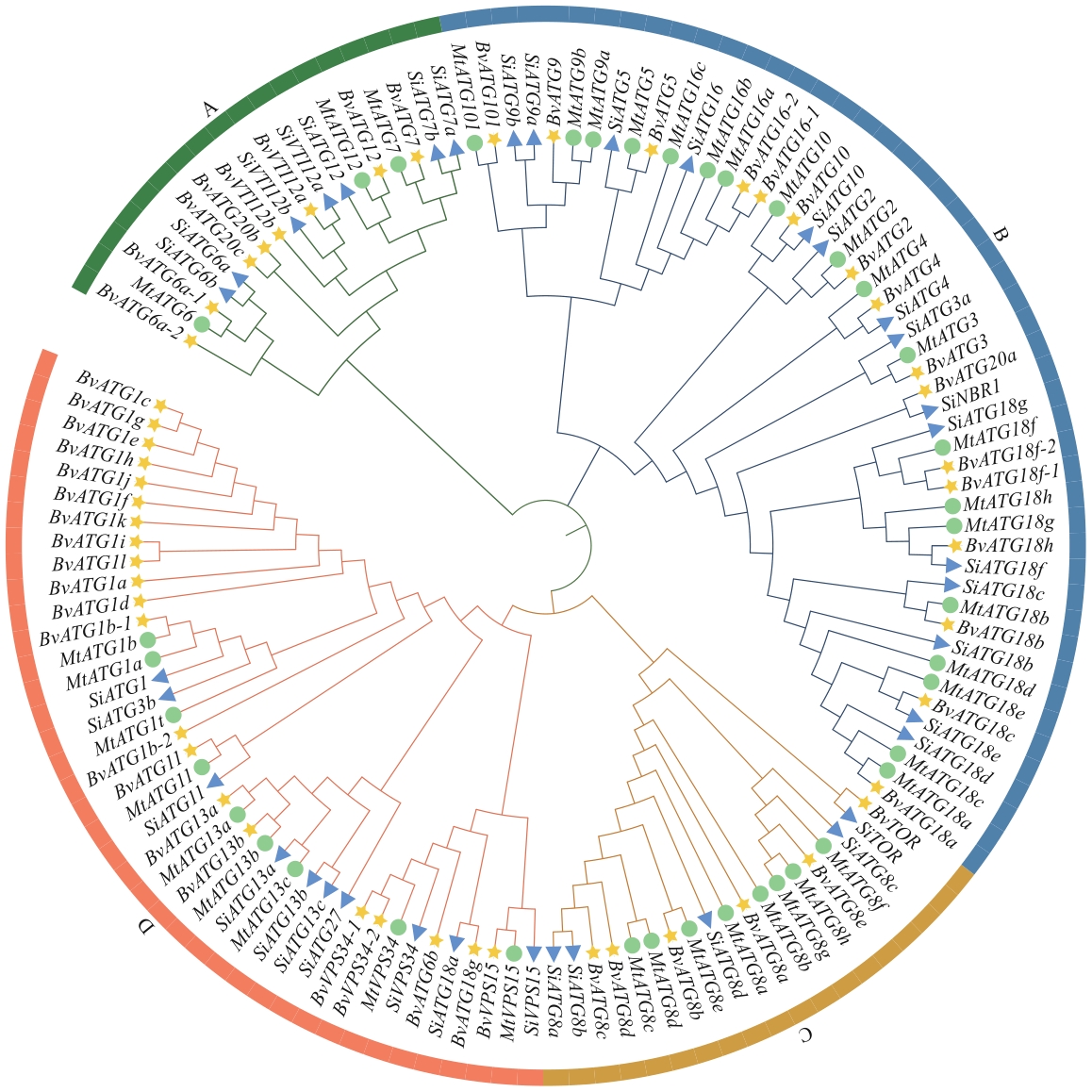
图2 高等植物ATGs系统进化分析黄色五角星代表甜菜,绿色圆圈表示苜蓿,蓝色三角形表示谷子;ATGs基因名称和登录号见附表2
Fig. 2 Phylogenetic analysis of ATGs in higher plantsThe yellow pentagons indicate sugar beet (B. vulgaris), the green circles indicate M. truncatula and the blue triangles indicate S. italica. The name and accession number of ATGs are shown in the Supplementary Table S2
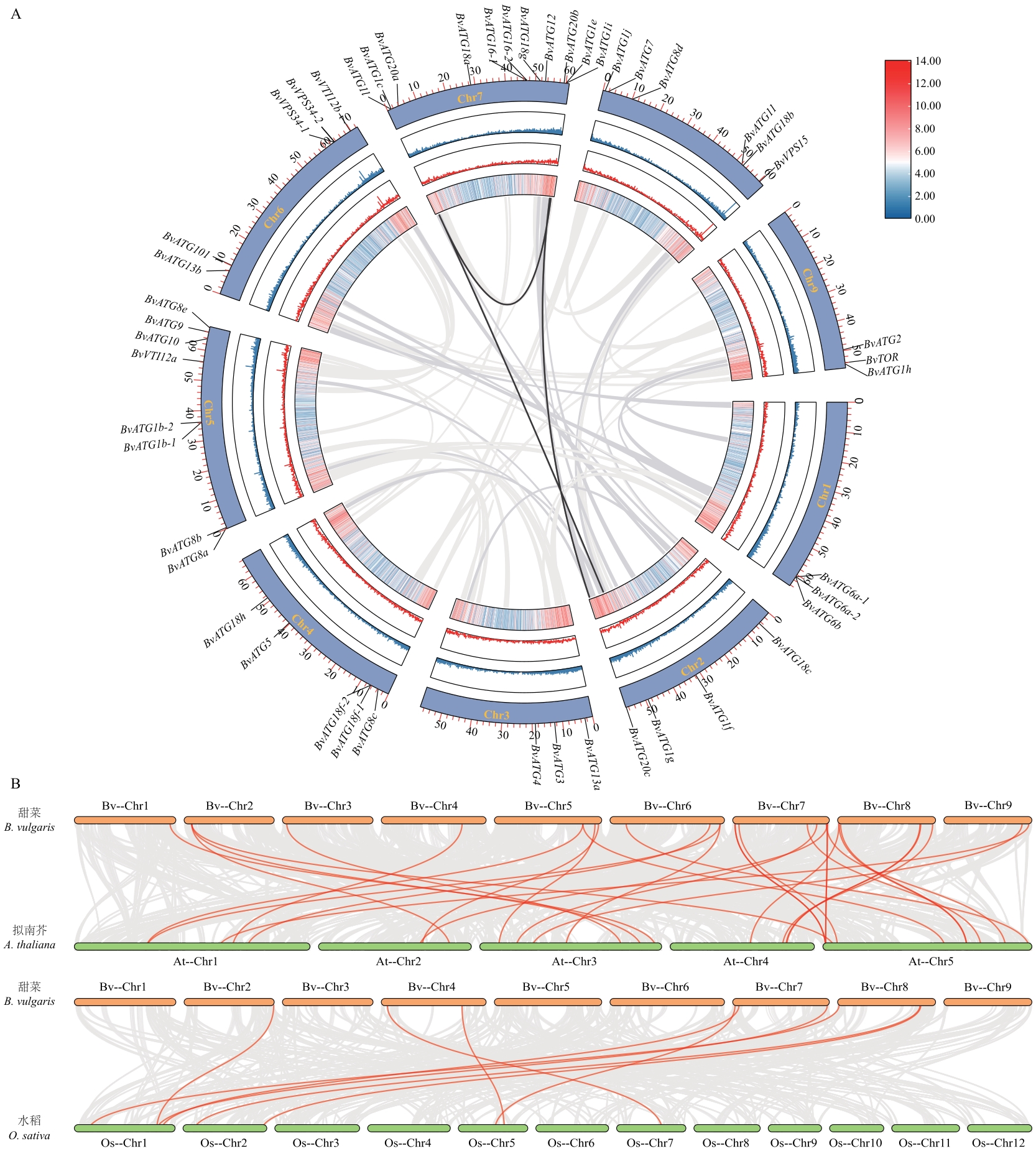
图5 甜菜BvATGs基因家族的共线性分析A:ATGs基因家族种内共线性分析;B:甜菜与拟南芥和水稻之间ATG基因的共线性分析
Fig. 5 Collinearity analysis of BvATGs gene family in sugar beetA: ATGs gene family intraspecies collinearity analysis. B: Synteny analysis of ATG genes between sugar beet (B. vulgaris), rice (O. sativa) and A. thaliana

图6 NaCl处理12 h (A)和24 h (B)的甜菜叶片和根中BvATGs相对表达水平将数据标准化为BvACTIN表达水平。竖线表示标准误差(SE)(n = 3)。柱子上不同小写字母表示P < 0.05水平差异显著。下同
Fig. 6 Relative expressions of BvATGs in the shoots and roots of sugar beet plants treated with different concentrations of NaCl for 12 h (A) and 24 h (B)Data are normalized to BvACTIN expression. Vertical bars indicate standard error (SE) (n = 3). Different lowercase letters on the bars indicate significant difference at P < 0.05 level. The same below
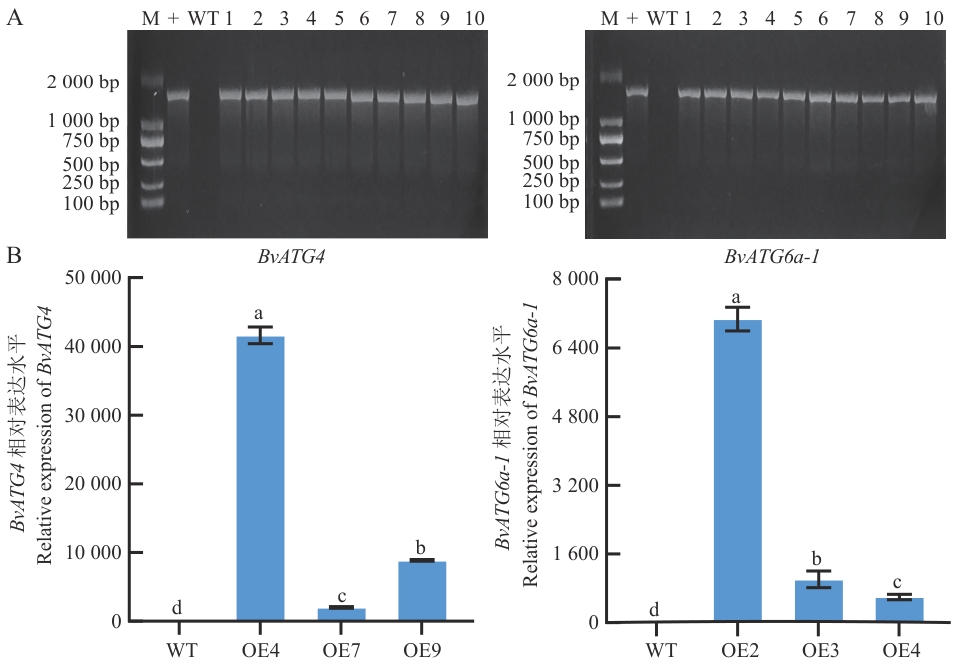
图8 BvATG4和BvATG6a-1转基因拟南芥鉴定A:BvATG4和BvATG6a-1转基因拟南芥基因组DNA PCR检测;B:BvATG4和BvATG6a-1转基因拟南芥RT-qPCR检测;M:DL2000分子量标记物;+:pEG-BvATG4、pEG-BvATG6a-1质粒;WT:野生型拟南芥;1‒10:BvATG4和BvATG6a-1转基因拟南芥;OE4、OE7、OE9:BvATG4转基因拟南芥;OE2、OE3、OE4:BvATG6a-1转基因拟南芥。将数据标准化为AtUBQ1表达水平
Fig. 8 Identification of BvATG4 and BvATG6a-1 transgenic A. thalianaA: Genomic DNA PCR detection of BvATG4 and BvATG6a-1 transgenic A. thaliana. B: RT-qPCR detection of BvATG4 and BvATG6a-1 transgenic A. thaliana. M: DL2000 marker for molecular weight; + : pEG-BvATG4 and pEG-BvATG6a-1 plasmid. WT: Wild-type A. thaliana; 1-10: BvATG4 and BvATG6a-1 transgenic A. thaliana; OE4, OE7, OE9: BvATG4 transgenic A. thaliana; OE2, OE3, OE4: BvATG6a-1 transgenic A. thaliana
| [1] | Liang XY, Li JF, Yang YQ, et al. Designing salt stress-resilient crops: Current progress and future challenges [J]. J Integr Plant Biol, 2024, 66(3): 303-329. |
| [2] | Zhou HP, Shi HF, Yang YQ, et al. Insights into plant salt stress signaling and tolerance [J]. J Genet Genom, 2024, 51(1): 16-34. |
| [3] | Ashraf M, Munns R. Evolution of approaches to increase the salt tolerance of crops [J]. Crit Rev Plant Sci, 2022, 41(2): 128-160. |
| [4] | Azmat MA, Zaheer M, Shaban M, et al. Autophagy: a new avenue and biochemical mechanisms to mitigate the climate change [J]. Scientifica, 2024, 2024(1): 9908323. |
| [5] | Yagyu M, Yoshimoto K. New insights into plant autophagy: molecular mechanisms and roles in development and stress responses [J]. J Exp Bot, 2024, 75(5): 1234-1251. |
| [6] | Sedaghatmehr M, Balazadeh S. Autophagy: a key player in the recovery of plants from heat stress [J]. J Exp Bot, 2024, 75(8): 2246-2255. |
| [7] | Suttangkakul A, Li FQ, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis [J]. Plant Cell, 2011, 23(10): 3761-3779. |
| [8] | Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae [J]. FEBS Lett, 1993, 333(1/2): 169-174. |
| [9] | Huang L, Wen X, Jin L, et al. HOOKLESS1 acetylates AUTOPHAGY-RELATED PROTEIN18a to promote autophagy during nutrient starvation in Arabidopsis [J]. Plant Cell, 2023, 36(1): 136-157. |
| [10] | Kwon SI, Park OK. Autophagy in plants [J]. J Plant Biol, 2008, 51(5): 313-320. |
| [11] | Han SJ, Yu BJ, Wang Y, et al. Role of plant autophagy in stress response [J]. Protein Cell, 2011, 2(10): 784-791. |
| [12] | Yadav M, Saxena G, Verma RK, et al. Genome-wide identification and expression analysis of autophagy-related genes (ATG) in Gossypium spp. reveals their crucial role in stress tolerance [J]. S Afr N J Bot, 2024, 167: 82-93. |
| [13] | Liu H, Liu Y, Zhang YT, et al. Genome-wide identification and expression analysis of autophagy-related genes (ATGs), revealing ATG8a and ATG18b participating in drought stress in Phoebe bournei [J]. Environ Exp Bot, 2024, 228: 106012. |
| [14] | Wang Y, Ban QY, Liu TJ, et al. Genome-wide identification and expression analysis of autophagy-related genes in eggplant (Solanum melongena L.) [J]. Sci Hortic, 2024, 330: 113085. |
| [15] | Wang Y, Sun X, Zhang ZW, et al. Genome-wide identification and characterization of the PbrATG family in Pyrus bretschneideri and functional analysis of PbrATG1a in response to Botryosphaeria dothidea [J]. Hortic Plant J, 2024, 10(2): 327-340. |
| [16] | Shi Y, Wu Y, Li ML, et al. Genome-wide identification and analysis of autophagy-related (ATG) genes in Lycium ruthenicum Murray reveals their crucial roles in salt stress tolerance [J]. Plant Sci, 2025, 352: 112371. |
| [17] | Luo KS, Li JH, Lu M, et al. Genome-wide identification and expression analysis of Rosa roxburghii autophagy-related genes in response to top-rot disease [J]. Biomolecules, 2023, 13(3): 556. |
| [18] | Petersen M, Avin-Wittenberg T, Bassham DC, et al. Autophagy in plants [J]. Autophagy Rep, 2024, 3(1): 2395731. |
| [19] | Luo LM, Zhang PP, Zhu RH, et al. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis [J]. Front Plant Sci, 2017, 8: 1459. |
| [20] | Liu ML, Ma L, Tang Y, et al. Maize autophagy-related protein ZmATG3 confers tolerance to multiple abiotic stresses [J]. Plants, 2024, 13(12): 1637. |
| [21] | Su WL, Bao Y, Lu YY, et al. Poplar autophagy receptor NBR1 enhances salt stress tolerance by regulating selective autophagy and antioxidant system [J]. Front Plant Sci, 2020, 11: 568411. |
| [22] | Yu XQ, Su WL, Zhang H, et al. Genome-wide analysis of autophagy-related gene family and PagATG18a enhances salt tolerance by regulating ROS homeostasis in poplar [J]. Int J Biol Macromol, 2023, 224: 1524-1540. |
| [23] | Yang J, Qiu LN, Mei QL, et al. MdHB7-like positively modulates apple salt tolerance by promoting autophagic activity and Na+ efflux [J]. Plant J, 2023, 116(3): 669-689. |
| [24] | Li NN, Wang W, Guo XT, et al. BvBZR1 improves parenchyma cell development and sucrose accumulation in sugar beet (Beta vulgaris L.) taproot [J]. Front Plant Sci, 2025, 16: 1495161. |
| [25] | Obama R, Kumagai K, Maruyama H, et al. Effect of different light conditions on the growth response of sugar beet to NaCl [J]. Soil Sci Plant Nutr, 2025, 71(2): 113-121. |
| [26] | Yang L, Ma CQ, Wang LL, et al. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14 [J]. J Plant Physiol, 2012, 169(9): 839-850. |
| [27] | Sánchez-Sastre LF, Alte da Veiga NMS, Ruiz-Potosme NM, et al. Sugar beet agronomic performance evolution in NW Spain in future scenarios of climate change [J]. Agronomy, 2020, 10(1): 91. |
| [28] | Lin XC, Song BQ, Adil MF, et al. Response of the rhizospheric soil microbial community of sugar beet to nitrogen application: a case of black soil in Northeast China [J]. Appl Soil Ecol, 2023, 191: 105050. |
| [29] | Wu GQ, Feng RJ, Liang N, et al. Sodium chloride stimulates growth and alleviates sorbitol-induced osmotic stress in sugar beet seedlings [J]. Plant Growth Regul, 2015, 75(1): 307-316. |
| [30] | Dohm JC, Minoche AE, Holtgräwe D, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris) [J]. Nature, 2014, 505(7484): 546-549. |
| [31] | Hu YD, Ren PP, Wei M, et al. Genome-wide identification of shaker K+ channel gene family in sugar beet (Beta vulgaris L.) and function of BvSKOR in response to salt and drought stresses [J]. Environ Exp Bot, 2024, 228: 106034. |
| [32] | Zhang JL, Wu GQ, Ma BT, et al. Genome-wide identification of the BvHSFs gene family and their expression in sugar beet (Beta vulgaris L.) under salt stress [J]. Sugar Tech, 2024, 26(5): 1463-1476. |
| [33] | Zhang XM, Wu GQ, Wei M. Genome-wide identification of sugar beet (Beta vulgaris L.) MAPKKKs gene family and their expression in response to salt stress [J]. Sugar Tech, 2024, 26(5): 1337-1349. |
| [34] | Ionascu A, Al Ecovoiu A, Chifiriuc MC, et al. qDATA - an R application implementing a practical framework for analyzing quantitative real-time PCR data [J]. Biotechniques, 2024, 76(12): 559-573. |
| [35] | Yang MK, Wang LP, Chen CM, et al. Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress [J]. Sci Rep, 2021, 11(1): 22933. |
| [36] | Taneja M, Tyagi S, Sharma S, et al. Ca2+ cation antiporters (CaCA): identification, characterization and expression profiling in bread wheat (Triticum aestivum L.) [J]. Front Plant Sci, 2016, 7: 1775. |
| [37] | Hu YH, Lehrach H, Janitz M. Comparative analysis of an experimental subcellular protein localization assay and in silico prediction methods [J]. J Mol Histol, 2009, 40(5/6): 343-352. |
| [38] | Qiao X, Li QH, Yin H, et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants [J]. Genome Biol, 2019, 20(1): 38. |
| [39] | Qiao X, Yin H, Li LT, et al. Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri) [J]. Front Plant Sci, 2018, 9: 161. |
| [40] | Hu YF, Zhang M, Yin FR, et al. Genome-wide identification and expression analysis of BrATGs and their different roles in response to abiotic stresses in Chinese cabbage [J]. Agronomy, 2022, 12(12): 2976. |
| [41] | Sheshadri SA, Nishanth MJ, Simon B. Stress-mediated cis-element transcription factor interactions interconnecting primary and specialized metabolism in planta [J]. Front Plant Sci, 2016, 7: 1725. |
| [42] | Wu XY, Xia M, Su P, et al. MYB transcription factors in plants: a comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism [J]. Int J Biol Macromol, 2024, 282(Pt 2): 136652. |
| [43] | Dong P, Xiong FJ, Que YM, et al. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis [J]. Front Plant Sci, 2015, 6: 677. |
| [44] | Fang Y, Wang S, Wu HL, et al. Genome-wide identification of ATG gene family members in Fagopyrum tataricum and their expression during stress responses [J]. Int J Mol Sci, 2022, 23(23): 14845. |
| [45] | Zhai YF, Guo M, Wang H, et al. Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L [J]. Front Plant Sci, 2016, 7: 131. |
| [46] | Fu XZ, Zhou X, Xu YY, et al. Comprehensive analysis of autophagy-related genes in sweet orange (Citrus sinensis) highlights their roles in response to abiotic stresses [J]. Int J Mol Sci, 2020, 21(8): 2699. |
| [47] | Wang JR, Zhou GQ, Huang WX, et al. Autophagy-related gene PlATG6a is involved in mycelial growth, asexual reproduction and tolerance to salt and oxidative stresses in Peronophythora litchii [J]. Int J Mol Sci, 2022, 23(3): 1839. |
| [1] | 曾厅, 张兰, 罗睿. 转录因子MpR2R3-MYB17调控地钱胞芽发育的功能研究[J]. 生物技术通报, 2026, 42(1): 208-217. |
| [2] | 康恺, 杨微, 李迎春, 谢为天, 吴海燕, 尤育品, 陈志宝. 桦褐孔菌醇通过激活Nrf2/PGC-1α/线粒体自噬防治AFB1诱导的小鼠肝损伤[J]. 生物技术通报, 2026, 42(1): 338-351. |
| [3] | 吕呈聪, 衡蒙, 陈思琪, 金雪花. 彩色马蹄莲花青素苷转运相关ZhGSTF的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 161-169. |
| [4] | 杨丹, 靳雅荣, 毛春力, 王碧娴, 张雅宁, 杨智怡, 周芷瑶, 杨锐鸣, 范恒睿, 黄琳凯, 严海东. 象草C2H2基因家族鉴定及表达分析[J]. 生物技术通报, 2026, 42(1): 251-261. |
| [5] | 杨跃琴, 邢英, 仲子荷, 田维军, 杨雪清, 王建旭. 甲基汞胁迫下水稻OsMATE34的表达及功能分析[J]. 生物技术通报, 2026, 42(1): 86-94. |
| [6] | 杨娟, 冯慧, 吉乃喆, 孙丽萍, 王赟, 张佳楠, 赵世伟. 月季AP2/ERF转录因子RcERF4和RcRAP2-12的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 150-160. |
| [7] | 张月, 戴月华, 张莹莹, 李奥辉, 李楚慧, 薛金爱, 秦慧彬, 陈妍, 聂萌恩, 张海平. 大豆烯酰辅酶A还原酶ECR14基因的克隆与功能分析[J]. 生物技术通报, 2026, 42(1): 95-104. |
| [8] | 吴翠翠, 陈登科, 兰刚, 夏芝, 李朋波. 花生转录因子AhHDZ70的生物信息学分析及耐盐耐旱性研究[J]. 生物技术通报, 2026, 42(1): 198-207. |
| [9] | 陈静欢, 房国楠, 朱文豪, 叶广继, 苏旺, 贺苗苗, 杨生龙, 周云. 马铃薯种质资源淀粉表征及相关基因表达分析[J]. 生物技术通报, 2026, 42(1): 170-183. |
| [10] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [11] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [12] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [13] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [14] | 黄国栋, 邓宇星, 程宏伟, 但焱南, 周会汶, 吴兰花. 大豆ZIP基因家族鉴定及响应铝胁迫的表达分析[J]. 生物技术通报, 2025, 41(9): 71-81. |
| [15] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||