








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 208-217.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0511
收稿日期:2025-05-18
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
罗睿,男,博士,副教授,研究方向 :植物学和遗传学;E-mail: rluo1@gzu.edu.cn作者简介:曾厅,女,硕士研究生,研究方向 :遗传学;E-mail: 1774837790@qq.com
基金资助:
ZENG Ting1,2,3( ), ZHANG Lan1,2, LUO Rui1(
), ZHANG Lan1,2, LUO Rui1( )
)
Received:2025-05-18
Published:2026-01-26
Online:2026-02-04
摘要:
目的 探究MpR2R3-MYB17基因在地钱胞芽发育调控中的功能及分子机制,为进一步揭示地钱胞芽发育调控机制提供新的依据。 方法 基于地钱基因组信息克隆MpR2R3-MYB17的全长CDS序列,对其编码蛋白质进行生物信息学分析并瞬时转化烟草检测MpR2R3-MYB17亚细胞定位。构建过表达载体和CRISPR/Cas9基因编辑载体,利用农杆菌介导的非组培依赖的地钱转化方法获得MpR2R3-MYB17过表达株系和突变体株系以评估表型变化。 结果 生物信息学分析表明,MpR2R3-MYB17的开放阅读框的长度为1 277 bp,编码421个氨基酸,具有2个保守的SANT序列,是R2R3型MYB转录因子。系统发育树揭示MpR2R3-MYB17与拟南芥R2R3-MYB第9亚家族聚为一支,其中与AtMYB17同源性最高。亚细胞定位分析表明,MpR2R3-MYB17蛋白定位于细胞核内,提示其可能直接参与转录调控过程。表型分析表明,与野生型相比,过表达株系的叶状体上胞芽杯密度和胞芽数量显著增加,或在高表达状态下形成未分化细胞团,且叶状体的生长受到抑制。突变体株系的胞芽杯密度及单个胞芽杯内胞芽数量均较野生型显著降低。 结论 MpR2R3-MYB17作为细胞核定位的转录因子参与调控地钱胞芽杯和胞芽的发育过程。
曾厅, 张兰, 罗睿. 转录因子MpR2R3-MYB17调控地钱胞芽发育的功能研究[J]. 生物技术通报, 2026, 42(1): 208-217.
ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L.[J]. Biotechnology Bulletin, 2026, 42(1): 208-217.
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Usage |
|---|---|---|
| M-F | GCTCTAGAATGGGGAGAGCGCCTTG | 基因克隆 |
| M-R | GCCCCCGGGTCAGCGCTCCATGACGGC | |
| gRT1 | TGCTGCGATAAGCAGGGGTTgttttagagctagaaat | sgRNA表达盒构建 |
| gRT2 | CGGGCACTTCCAAAACATGCgttttagagctagaaat | |
| gRNA-R | CGGAGGAAAATTCCATCCAC | |
| P-F | AGCGTGggtctcGCTCGACGCGTATCCATCCACTCCAAGCTC | |
| P-R | TTCAGAggtctcTACCGACTAGTATGGAATCGGCAGCAAAGG | |
| GUS-F | TCTGCGACGCTCACACCGAT | 过表达阳性植株鉴定 |
| GUS-R | GCCAACGCGCAATATGCCTT | |
| qMpR2R3-MYB17-F | CTACTGGAACACGCACCTGA | 实时荧光定量PCR |
| qMpR2R3-MYB17-R | AGATGACTCAACTTGGAGCACA | |
| qMpAPT-F | CGAAAGCCCAAGAAGCTACC | 内参基因 |
| qMpAPT-R | GTACCCCCGGTTGCAATAAG | |
| Hyg-F | GTCGTCCATCACAGTTTGCC | 突变体阳性鉴定 |
| Hyg-R | TGCTTGACATTGGGGAGTTT | |
| PegMYB17-F | TATATTTGCCGTTGCTGCCG | 测序片段克隆 |
| PegMYB17-R | AGGGTTCAAAGCGCGATTCA |
表1 本研究使用的引物
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Usage |
|---|---|---|
| M-F | GCTCTAGAATGGGGAGAGCGCCTTG | 基因克隆 |
| M-R | GCCCCCGGGTCAGCGCTCCATGACGGC | |
| gRT1 | TGCTGCGATAAGCAGGGGTTgttttagagctagaaat | sgRNA表达盒构建 |
| gRT2 | CGGGCACTTCCAAAACATGCgttttagagctagaaat | |
| gRNA-R | CGGAGGAAAATTCCATCCAC | |
| P-F | AGCGTGggtctcGCTCGACGCGTATCCATCCACTCCAAGCTC | |
| P-R | TTCAGAggtctcTACCGACTAGTATGGAATCGGCAGCAAAGG | |
| GUS-F | TCTGCGACGCTCACACCGAT | 过表达阳性植株鉴定 |
| GUS-R | GCCAACGCGCAATATGCCTT | |
| qMpR2R3-MYB17-F | CTACTGGAACACGCACCTGA | 实时荧光定量PCR |
| qMpR2R3-MYB17-R | AGATGACTCAACTTGGAGCACA | |
| qMpAPT-F | CGAAAGCCCAAGAAGCTACC | 内参基因 |
| qMpAPT-R | GTACCCCCGGTTGCAATAAG | |
| Hyg-F | GTCGTCCATCACAGTTTGCC | 突变体阳性鉴定 |
| Hyg-R | TGCTTGACATTGGGGAGTTT | |
| PegMYB17-F | TATATTTGCCGTTGCTGCCG | 测序片段克隆 |
| PegMYB17-R | AGGGTTCAAAGCGCGATTCA |
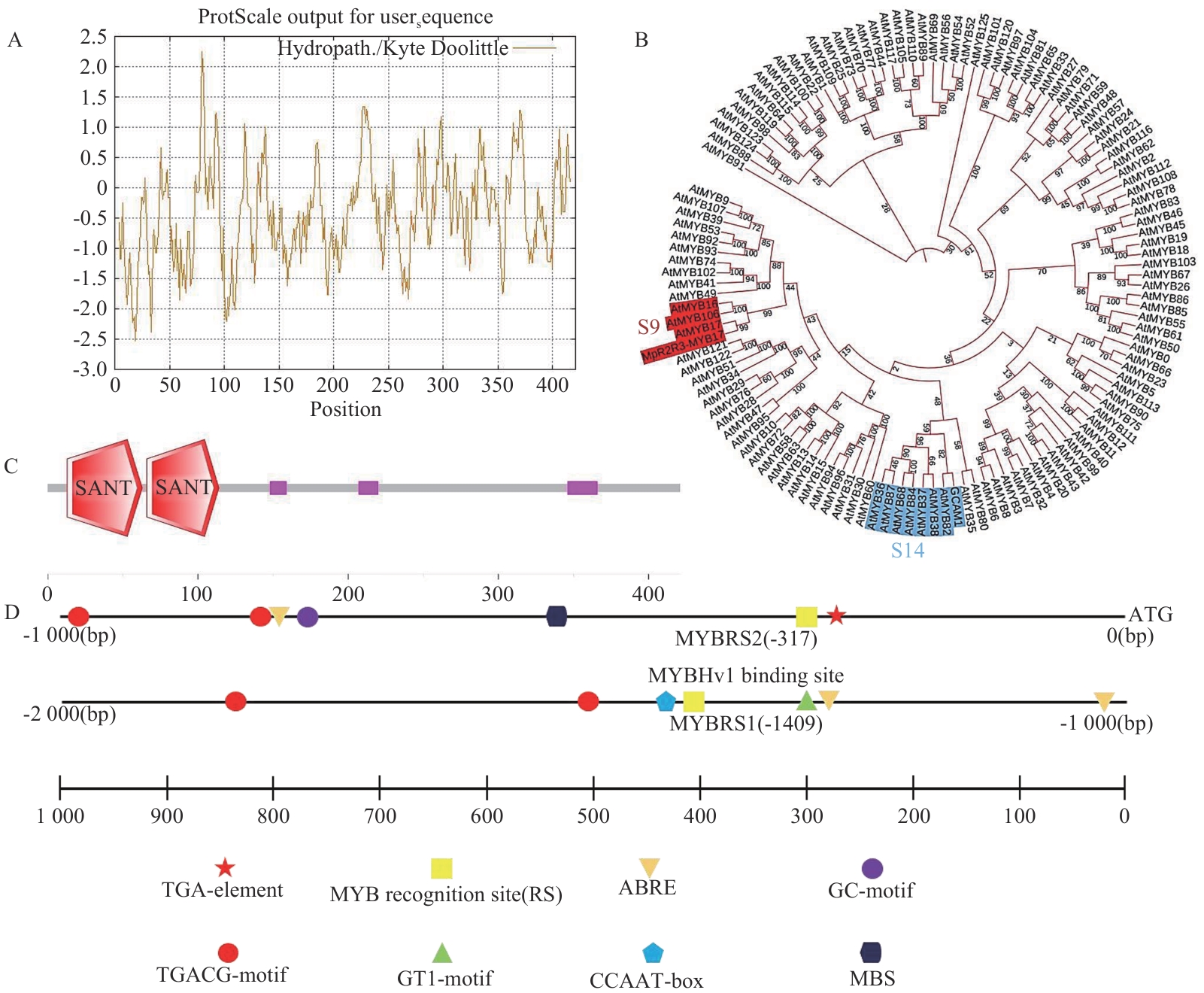
图1 生物信息学分析和系统发育树A:MpR2R3-MYB17蛋白氨基酸序列亲疏水性预测结果;B:系统发育树;C:MpR2R3-MYB17蛋白保守结构域分析(红色部分:SANT结构域;紫色部分:低复杂度区域);D:MpR2R3-MYB17启动子区顺式作用元件分析
Fig. 1 Bioinformatics analysis and phylogenetic treeA: Hydrophilicity/hydrophobicity prediction of theMpR2R3-MYB17 protein amino acid sequence. B: Phylogenetic tree. C: Conserved domain analysis of the MpR2R3-MYB17 protein (Red: SANT domain. Purple: Low-complexity region). D: Analysis of cis-regulatory elements in the MpR2R3-MYB17 gene promoter region
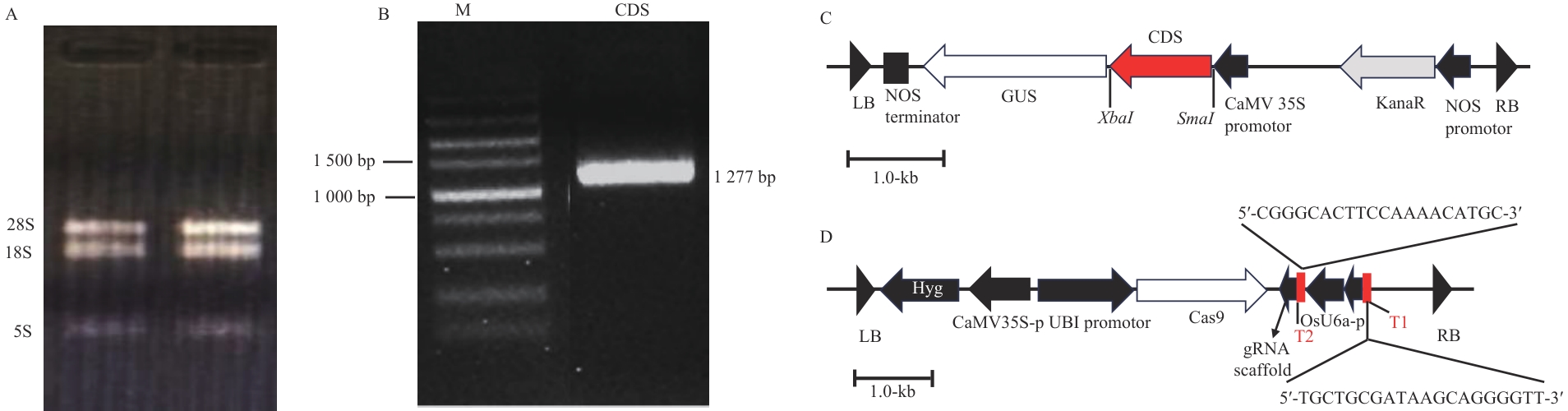
图2 RNA提取、MpR2R3-MYB17基因克隆、重组质粒图A:RNA提取结果;B:MpR2R3-MYB17基因克隆,产物长度为1 277 bp;C:pBI121-MpR2R3-MYB17重组质粒图(GUS:标记基因);D:pEGCas9Pubi-MpR2R3-MYB17重组质粒(T1、T2为sgRNA靶点序列;gRNA scaffold为gRNA骨架区)
Fig. 2 RNA extraction, cloning of MpR2R3-MYB17 gene, and recombinant plasmid mapsA: RNA extraction. B: MpR2R3-MYB17 gene cloning, the product length is 1 277 bp. C: pBI121-MpR2R3-MYB17 recombinant plasmid map (labelled gene). D: pEGCas9Pubi-MpR2R3-MYB17 recombinant plasmid (T1 and T2: sgRNA target sequences. gRNA scaffold: gRNA structural backbone)

图4 抗性植株筛选、过表达植株PCR鉴定和GUS染色A:农杆菌侵染处理后的地钱胞芽;B:经Kana抗性筛选获得的抗性植株;C:MpR2R3-MYB17过表达转基因植株的PCR阳性鉴定;D:MpR2R3-MYB17过表达植株的组织化学GUS染色
Fig. 4 Screening of resistant plants, PCR identification of overexpressed plants and GUS stainingA: Marchantia polymorpha gemmae following Agrobacterium tumefaciens infection. B: Kanamycin-resistant plants selected after transformation. C: PCR confirmation of MpR2R3-MYB17-overexpressed transgenic lines. D: Histochemical GUS staining in MpR2R3-MYB17-overexpressed plants
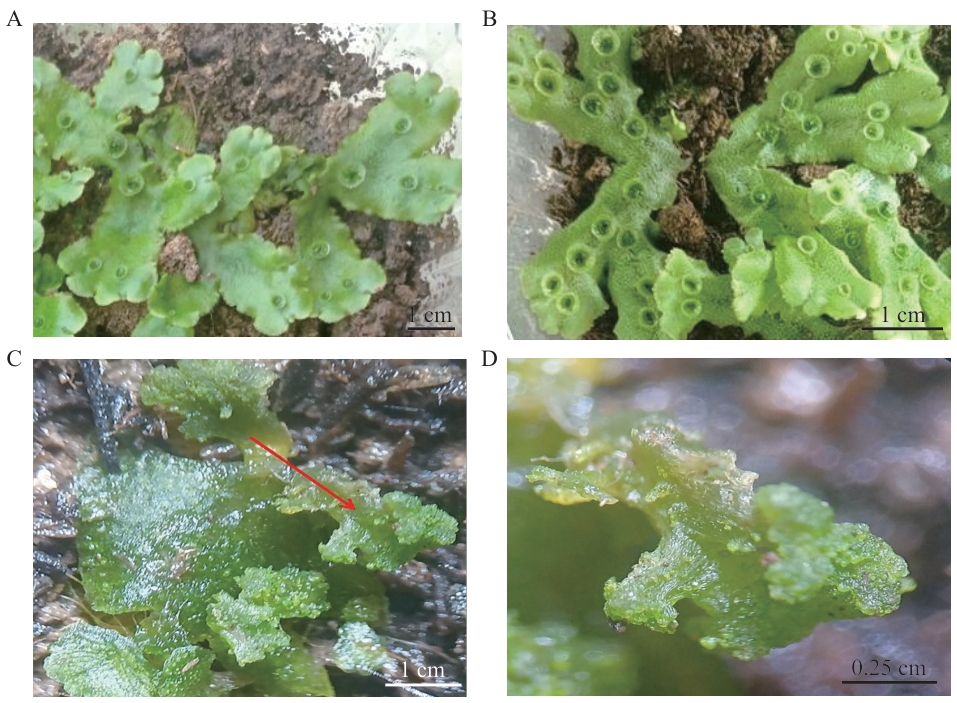
图5 过表达植株表型A:地钱野生型植株;B:MpR2R3-MYB17过表达株系表型(胞芽杯密度增加);C:MpR2R3-MYB17过表达株系表型(产生未分化细胞团),红色箭头表示未分化细胞团;D:图C中未分化细胞团的放大图
Fig. 5 Phenotype of overexpressing plantsA: Wild type of M. polymorpha L.. B: Phenotypes of overexpressing plant lines (increased gemma cup density). C: Phenotypes of overexpressing plant lines (undifferentiated cell clumps). Undifferentiated cell clumps are indicated by red arrows. D: Enlarged view of undifferentiated cell clumps from panel C
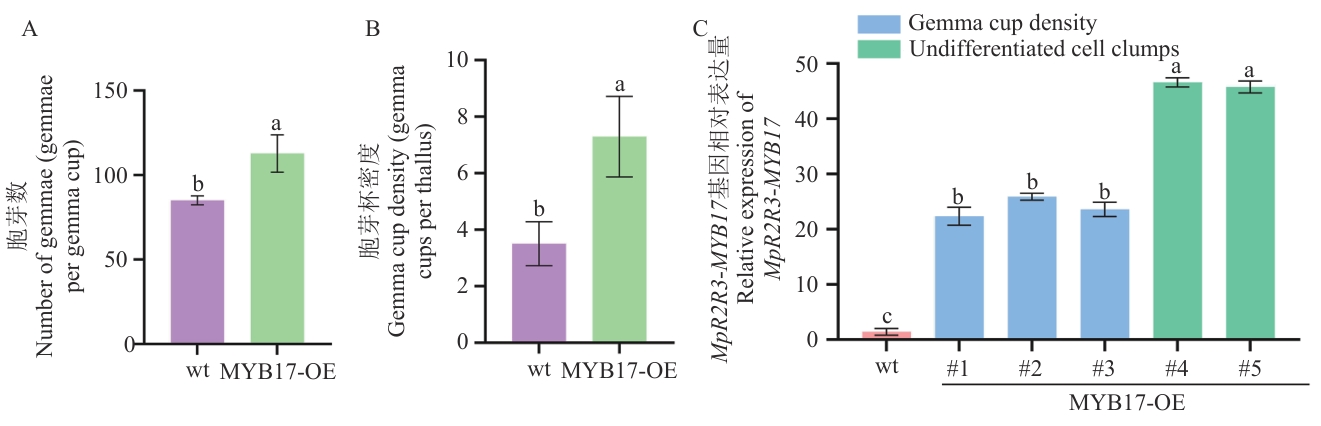
图6 过表达株系表型统计与表达分析A:单个胞芽杯内胞芽数量(个/胞芽杯);B:胞芽杯密度(个/叶状体);C:MpR2R3-MYB17基因相对表达量(wt:野生型;MYB17-OE#1/2/3:胞芽杯数量增多的过表达株系;MYB17-OE#4/5:产生未分化细胞团的过表达株系);不同小写字母表示组间差异显著(P<0.05)
Fig. 6 Phenotypic statistics and expression analysis of overexpressed plant linesA: Number of gemmae per gemma cup (gemmae/gemma cup). B: Gemma cup density (gemma cups per thallus). C: Relative expression of MpR2R3-MYB17 (wt: wild-type. MYB17-OE#1/2/3: Overexpressed lines with increased gemma cup density. MYB17-OE#4/5: Overexpressed lines producing undifferentiated cell clumps). Different lowercase letters indicate significant differences between groups (P<0.05)
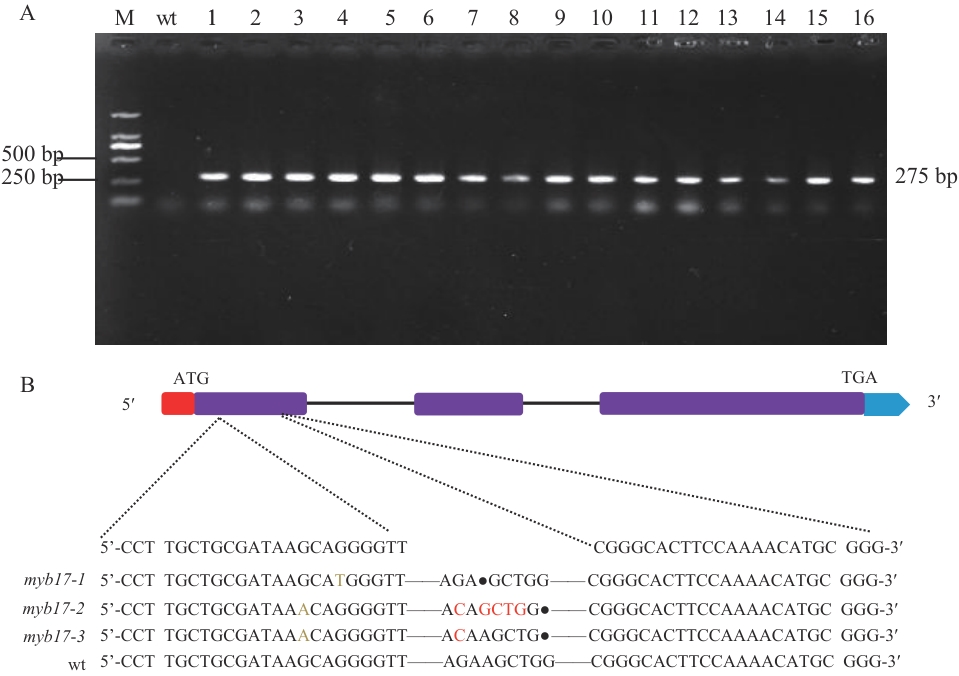
图7 突变体鉴定与序列比对A:转基因植株PCR鉴定;B:突变体序列比对(红色方框:5′非编码区;紫色方框:外显子区域;黑色横线:内含子区域;蓝色方框:3′非编码区;序列比对中红色字母表示碱基替换;圆点“·”表示碱基缺失;横线“—”表示与参考序列一致而被省略的未突变碱基)
Fig. 7 Mutant identification and sequence alignmentA: PCR identification of transgenic plants. B: Mutant sequence alignment (red box: 5′ non-coding region; purple boxes: exons; blue box: 3′ non-coding region. In the sequence alignment, red letters denote nucleotide substitutions. Dots "·": Base deletions. Horizontal line "—": Unmutated bases that are consistent with the reference sequence and are omitted)
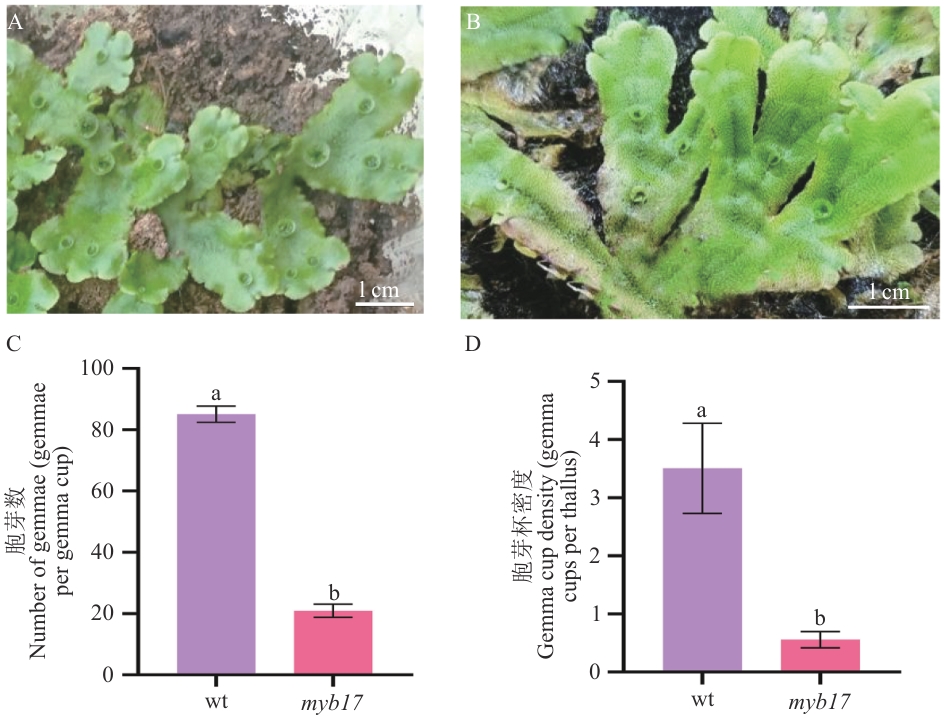
图8 突变体表型分析A:野生型植株;B:突变体株系;C:单个胞芽杯内胞芽数量(个/胞芽杯);D:胞芽杯密度(个/叶状体)。myb17代表3个独立突变体株系myb17-1、myb17-2、myb17-3的表型统计结果,不同小写字母表示组间差异显著(P<0.01)
Fig. 8 Phenotypic analysis of mutantsA: Wild type of M. polymorpha L.. B:Mutant lines. C:Number of gemmae per gemma cup (gemmae/gemma cup). D: Gemma cup density (gemma cups per thallus). myb17 indicates the phenotypic statistics of three independent mutant lines, myb17-1, myb17-2 and myb17-3, and different lowercase letters indicate significant differences between groups (P<0.01)
| [1] | Beaulieu C, Libourel C, Mbadinga Zamar DL, et al. The Marchantia polymorpha pangenome reveals ancient mechanisms of plant adaptation to the environment [J]. Nat Genet, 2025, 57(3): 729-740. |
| [2] | Shimamura M. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system [J]. Plant Cell Physiol, 2016, 57(2): 230-256. |
| [3] | Kato H, Yasui Y, Ishizaki K. Gemma cup and gemma development in Marchantia polymorpha [J]. New Phytol, 2020, 228(2): 459-465. |
| [4] | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. Roles of MYB transcription factor in regulating the responses of plants to stress [J]. Biotechnol Bull, 2024, 40(6): 5-22. | |
| [5] | Zhang DW, Zhou HP, Zhang Y, et al. Diverse roles of MYB transcription factors in plants [J]. J Integr Plant Biol, 2025, 67(3): 539-562. |
| [6] | Lal M, Bhardwaj E, Chahar N, et al. Comprehensive analysis of 1R-and 2R-MYBs reveals novel genic and protein features, complex organisation, selective expansion and insights into evolutionary tendencies [J]. Funct Integr Genomics, 2022, 22(3): 371-405. |
| [7] | Yang JH, Zhang BH, Gu G, et al. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.) [J]. BMC Genomics, 2022, 23(1): 432. |
| [8] | Koo Y, Poethig RS. Expression pattern analysis of three R2R3-MYB transcription factors for the production of anthocyanin in different vegetative stages of Arabidopsis leaves [J]. Appl Biol Chem, 2021, 64(1): 5. |
| [9] | Qin W, Xie LH, Li YP, et al. An R2R3-MYB transcription factor positively regulates the glandular secretory trichome initiation in Artemisia annua L [J]. Front Plant Sci, 2021, 12: 657156. |
| [10] | Okumura T, Nomoto Y, Kobayashi K, et al. MYB3R-mediated active repression of cell cycle and growth under salt stress in Arabidopsis thaliana [J]. J Plant Res, 2021, 134(2): 261-277. |
| [11] | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| [12] | Preston J, Wheeler J, Heazlewood J, et al. AtMYB32 is required for normal pollen development in Arabidopsis thaliana [J]. Plant J, 2004, 40(6): 979-995. |
| [13] | Steiner-Lange S, Unte US, Eckstein L, et al. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers [J]. Plant J, 2003, 34(4): 519-528. |
| [14] | Yasui Y, Tsukamoto S, Sugaya T, et al. GEMMA CUP-ASSOCIATED MYB1, an ortholog of axillary meristem regulators, is essential in vegetative reproduction in Marchantia polymorpha [J]. Curr Biol, 2019, 29(23): 3987-3995.e5. |
| [15] | Sugano SS, Nishihama R. CRISPR/Cas9-based genome editing of transcription factor genes in Marchantia polymorpha [J]. Methods Mol Biol, 2018, 1830: 109-126. |
| [16] | Du YM, Liu YF, Hu JX, et al. CRISPR/Cas9 systems: Delivery technologies and biomedical applications [J]. Asian J Pharm Sci, 2023, 18(6): 100854. |
| [17] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [18] | Chen ZW, Halford NG, Liu CH. Real-time quantitative PCR primer design, reference gene selection, calculations and statistics [J]. Metabolites, 2023, 13(7): 806. |
| [19] | Maren NA, Duduit JR, Huang DB, et al. Stepwise optimization of real-time RT-PCR analysis [J]. Methods Mol Biol, 2023, 2653: 317-332. |
| [20] | Naramoto S, Hata Y, Fujita T, et al. The bryophytes Physcomitrium patens and Marchantia polymorpha as model systems for studying evolutionary cell and developmental biology in plants [J]. Plant Cell, 2022, 34(1): 228-246. |
| [21] | 关范圆, 郑语嫣, 米芯雨, 等. R2R3型MYB转录因子调控植物胁迫应答的研究进展 [J]. 植物医学, 2024, 3(3): 1-9. |
| Guan FY, Zheng YY, Mi XY, et al. Research advance of the functions of R2R3-MYB transcription factors in plant response to biotic and abiotic stresses [J]. Plant Health Med, 2024, 3(3): 1-9. | |
| [22] | 田琴, 刘奎, 吴翔纬, 等. 转录因子VcMYB17调控蓝莓抗旱性的功能研究 [J]. 生物技术通报, 2025, 41(4): 198-210. |
| Tian Q, Liu K, Wu XW, et al. Functional study of transcription factor VcMYB17 in regulating drought tolerance in blueberry [J]. Biotechnol Bull, 2025, 41(4): 198-210. | |
| [23] | Brockington SF, Alvarez-Fernandez R, Landis JB, et al. Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation [J]. Mol Biol Evol, 2013, 30(3): 526-540. |
| [24] | Jakoby MJ, Falkenhan D, Mader MT, et al. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106 [J]. Plant Physiol, 2008, 148(3): 1583-1602. |
| [25] | 张哲, 何鑫玺, 张纪利, 等. NtMYB17过表达对烟草腺毛发育的影响 [J]. 中国烟草科学, 2024, 45(3): 86-92. |
| Zhang Z, He XX, Zhang JL, et al. Effect of NtMYB17 overexpression on development of glandular trichomes in tobacco [J]. Chin Tob Sci, 2024, 45(3): 86-92. | |
| [26] | Machado A, Wu YR, Yang YM, et al. The MYB transcription factor GhMYB25 regulates early fibre and trichome development [J]. Plant J, 2009, 59(1): 52-62. |
| [27] | Tak H, Negi S, Ganapathi TR. Overexpression of MusaMYB31, a R2R3 type MYB transcription factor gene indicate its role as a negative regulator of lignin biosynthesis in banana [J]. PLoS One, 2017, 12(2): e0172695. |
| [28] | Proust H, Honkanen S, Jones VAS, et al. RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants [J]. Curr Biol, 2016, 26(1): 93-99. |
| [29] | Flores-Sandoval E, Eklund DM, Hong SF, et al. Class C ARFs evolved before the origin of land plants and antagonize differentiation and developmental transitions in Marchantia polymorpha [J]. New Phytol, 2018, 218(4): 1612-1630. |
| [30] | Aki SS, Mikami T, Naramoto S, et al. Cytokinin signaling is essential for organ formation in Marchantia polymorpha [J]. Plant Cell Physiol, 2019, 60(8): 1842-1854. |
| [1] | 龙林茜, 曾银萍, 王茜, 邓玉萍, 葛敏茜, 陈彦灼, 李鑫娟, 杨军, 邹建. 向日葵GH3基因家族鉴定及其在花发育中的功能分析[J]. 生物技术通报, 2026, 42(1): 125-138. |
| [2] | 任云儿, 伍国强, 成斌, 魏明. 甜菜BvATGs基因家族全基因组鉴定及盐胁迫下表达模式分析[J]. 生物技术通报, 2026, 42(1): 184-197. |
| [3] | 杨娟, 冯慧, 吉乃喆, 孙丽萍, 王赟, 张佳楠, 赵世伟. 月季AP2/ERF转录因子RcERF4和RcRAP2-12的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 150-160. |
| [4] | 张驰昊, 刘晋囡, 晁跃辉. 蒺藜苜蓿bZIP转录因子MtbZIP29的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 241-250. |
| [5] | 吴翠翠, 陈登科, 兰刚, 夏芝, 李朋波. 花生转录因子AhHDZ70的生物信息学分析及耐盐耐旱性研究[J]. 生物技术通报, 2026, 42(1): 198-207. |
| [6] | 陈强, 于璎霏, 张颖, 张冲. 茉莉酸甲酯对薄皮甜瓜‘绿宝石’采后冷害的调控[J]. 生物技术通报, 2025, 41(9): 105-114. |
| [7] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [8] | 史发超, 姜永华, 刘海伦, 文英杰, 严倩. 荔枝LcTFL1基因的克隆与功能分析[J]. 生物技术通报, 2025, 41(9): 159-167. |
| [9] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [10] | 王斌, 林冲, 袁晓, 蒋园园, 王玉昆, 肖艳辉. bHLH转录因子UNE10克隆及其在丁香罗勒挥发性化合物合成调控中的功能[J]. 生物技术通报, 2025, 41(9): 207-218. |
| [11] | 黄诗宇, 田姗姗, 杨天为, 高曼熔, 张尚文. 赤苍藤WRI1基因家族的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(8): 242-254. |
| [12] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [13] | 李雅琼, 格桑拉毛, 陈启迪, 杨宇环, 何花转, 赵耀飞. 异源过表达高粱SbSnRK2.1增强拟南芥对盐胁迫的抗性[J]. 生物技术通报, 2025, 41(8): 115-123. |
| [14] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [15] | 余永霞, 杜再慧, 朱龙佼, 许文涛. 基因编辑技术在牛种中的应用及研究进展[J]. 生物技术通报, 2025, 41(8): 34-41. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||