








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 50-65.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0627
郭发旭1,2( ), 冯全1(
), 冯全1( ), 张建华2,3(
), 张建华2,3( ), 周焕斌2,4(
), 周焕斌2,4( ), 杨森1, 王健2,3, 周国民5,6,7
), 杨森1, 王健2,3, 周国民5,6,7
收稿日期:2025-06-16
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
冯全,男,博士,教授,研究方向 :计算机视觉;E-mail: fquan@gsau.edu.cn作者简介:郭发旭,男,博士研究生,研究方向 :生物信息学;E-mail: guofax@gsau.edu.cn
基金资助:
GUO Fa-xu1,2( ), FENG Quan1(
), FENG Quan1( ), ZHANG Jian-hua2,3(
), ZHANG Jian-hua2,3( ), ZHOU Huan-bin2,4(
), ZHOU Huan-bin2,4( ), YANG Sen1, WANG Jian2,3, ZHOU Guo-min5,6,7
), YANG Sen1, WANG Jian2,3, ZHOU Guo-min5,6,7
Received:2025-06-16
Published:2025-12-26
Online:2026-01-06
摘要:
酶在生物体内以及工业应用里有着极其关键的作用,凭借其独特的催化性能,成为催化剂的关键选择之一。然而传统的酶改造与设计方法面临不少挑战,如序列空间庞大以及多目标优化存在困难等情况。近年来,人工智能(artificial intelligence, AI)技术中的深度学习和生成式人工智能方法,为酶改造与设计提供了新思路和解决方案,可在大规模数据的支撑下突破这些限制。AI驱动的策略实现了高效的序列空间探索和精准的结构-功能关系预测,并借助强化学习框架协调多目标优化。这些方法不仅显著加速了酶的工程化进程,更在提升催化效率、增强热稳定性、改善底物选择性等方面取得了突破性成果。本文系统综述了人工智能驱动酶改造与设计的最新研究进展,从基础数据库构建、智能改造策略、智能设计方法等方面进行了深入分析,同时探讨了当前面临的数据、模型及工程化挑战与未来发展方向。这些创新为设计高性能、多功能的酶开辟了广阔道路,并将推动生物制造、环境修复和生物育种等领域向更高效、智能和可持续的方向发展。
郭发旭, 冯全, 张建华, 周焕斌, 杨森, 王健, 周国民. 人工智能驱动的酶改造与设计研究进展[J]. 生物技术通报, 2025, 41(12): 50-65.
GUO Fa-xu, FENG Quan, ZHANG Jian-hua, ZHOU Huan-bin, YANG Sen, WANG Jian, ZHOU Guo-min. Research Advances in AI-driven Enzyme Modifying and Design[J]. Biotechnology Bulletin, 2025, 41(12): 50-65.

图1 AI驱动的酶改造与设计的要素和结构A:数据,从数据库中获得的数据;B:编码方式,从序列、结构、网络等中提取特征信息,分别为序列特征、结构特征和嵌入特征;C:建模,使用深度学习模型和蛋白质语言模型挖掘不同特征;D:测试,通过高通量测试平台进一步测试设计,或通过定向进化优化设计
Fig. 1 Components and framework of AI-driven enzyme engineering and designA: Data, acquiring data from databases. B: Encoding, extracting feature from sequence, structure, and network representations, including sequence feature, structural feature, and embedding feature. C: Modeling, applying deep learning models and protein language models to analyze and learn from the extracted features. D: Testing, experimental validation of designed enzymes using high-throughput screening platforms or further optimization via directed evolution
名称 Name | 类型 Type | 数目/大小 Number/Size | 特征 Features | 参考文献 Reference |
|---|---|---|---|---|
| UniProtKB | 蛋白质序列与功能注释 | >2亿蛋白质序列 | 高质量注释,Swiss-Prot与TrEMBL,广泛交叉引用 | [ |
| PDB / RCSB | 蛋白质三维结构 | >210 000结构条目 | 三维结构,支持多种解析方法,结构视图与功能注释 | [ |
| ProThermDB | 热力学稳定性(实验数据) | 25 000突变数据 | 含ΔΔG、Tm等稳定性参数,适用于建模和蛋白质设计 | [ |
| FireProtDB | 蛋白质突变稳定性(实验+预测) | >10 000突变 | 包含ΔΔG预测,适用于稳定性预测工具开发 | [ |
| SoluProtMutDB | 可溶性突变实验数据库 | >10 000突变 | 聚焦蛋白表达产物可溶性,支持可溶性优化 | [ |
| ProtaBank | 蛋白质工程实验数据 | >100 000数据条目 | 包含结合力、催化活性、稳定性等,支持上传与机器学习训练 | [ |
| AlphaFold DB | 蛋白质结构预测 | >2亿预测结构 | 基于深度学习,提供可信度评分,广泛补充实验结构缺口 | [ |
| GotEnzymes | 酶催化参数预测(AI生成) | >10亿酶-底物对 | 基于AI预测kcat,适用于合成生物学与模型代谢网络 | [ |
| InterPro | 蛋白结构域和家族分类 | >47 000结构域类型 | 综合Pfam等数据库,支持功能位点注释与家族进化研究 | [ |
| BRENDA | 酶功能与生化参数数据库 | >10万酶,>100万参数 | 涵盖Km、kcat、pH、温度、抑制剂等,按EC号系统整理 | [ |
| BKMS-react | 生化反应整合数据库(代谢建模) | >81 200酶催化反应 | 整合KEGG/BRENDA等,支持反应建模、底物产物分析 | [ |
表1 常见数据库汇总
Table 1 Summary of commonly used databases
名称 Name | 类型 Type | 数目/大小 Number/Size | 特征 Features | 参考文献 Reference |
|---|---|---|---|---|
| UniProtKB | 蛋白质序列与功能注释 | >2亿蛋白质序列 | 高质量注释,Swiss-Prot与TrEMBL,广泛交叉引用 | [ |
| PDB / RCSB | 蛋白质三维结构 | >210 000结构条目 | 三维结构,支持多种解析方法,结构视图与功能注释 | [ |
| ProThermDB | 热力学稳定性(实验数据) | 25 000突变数据 | 含ΔΔG、Tm等稳定性参数,适用于建模和蛋白质设计 | [ |
| FireProtDB | 蛋白质突变稳定性(实验+预测) | >10 000突变 | 包含ΔΔG预测,适用于稳定性预测工具开发 | [ |
| SoluProtMutDB | 可溶性突变实验数据库 | >10 000突变 | 聚焦蛋白表达产物可溶性,支持可溶性优化 | [ |
| ProtaBank | 蛋白质工程实验数据 | >100 000数据条目 | 包含结合力、催化活性、稳定性等,支持上传与机器学习训练 | [ |
| AlphaFold DB | 蛋白质结构预测 | >2亿预测结构 | 基于深度学习,提供可信度评分,广泛补充实验结构缺口 | [ |
| GotEnzymes | 酶催化参数预测(AI生成) | >10亿酶-底物对 | 基于AI预测kcat,适用于合成生物学与模型代谢网络 | [ |
| InterPro | 蛋白结构域和家族分类 | >47 000结构域类型 | 综合Pfam等数据库,支持功能位点注释与家族进化研究 | [ |
| BRENDA | 酶功能与生化参数数据库 | >10万酶,>100万参数 | 涵盖Km、kcat、pH、温度、抑制剂等,按EC号系统整理 | [ |
| BKMS-react | 生化反应整合数据库(代谢建模) | >81 200酶催化反应 | 整合KEGG/BRENDA等,支持反应建模、底物产物分析 | [ |
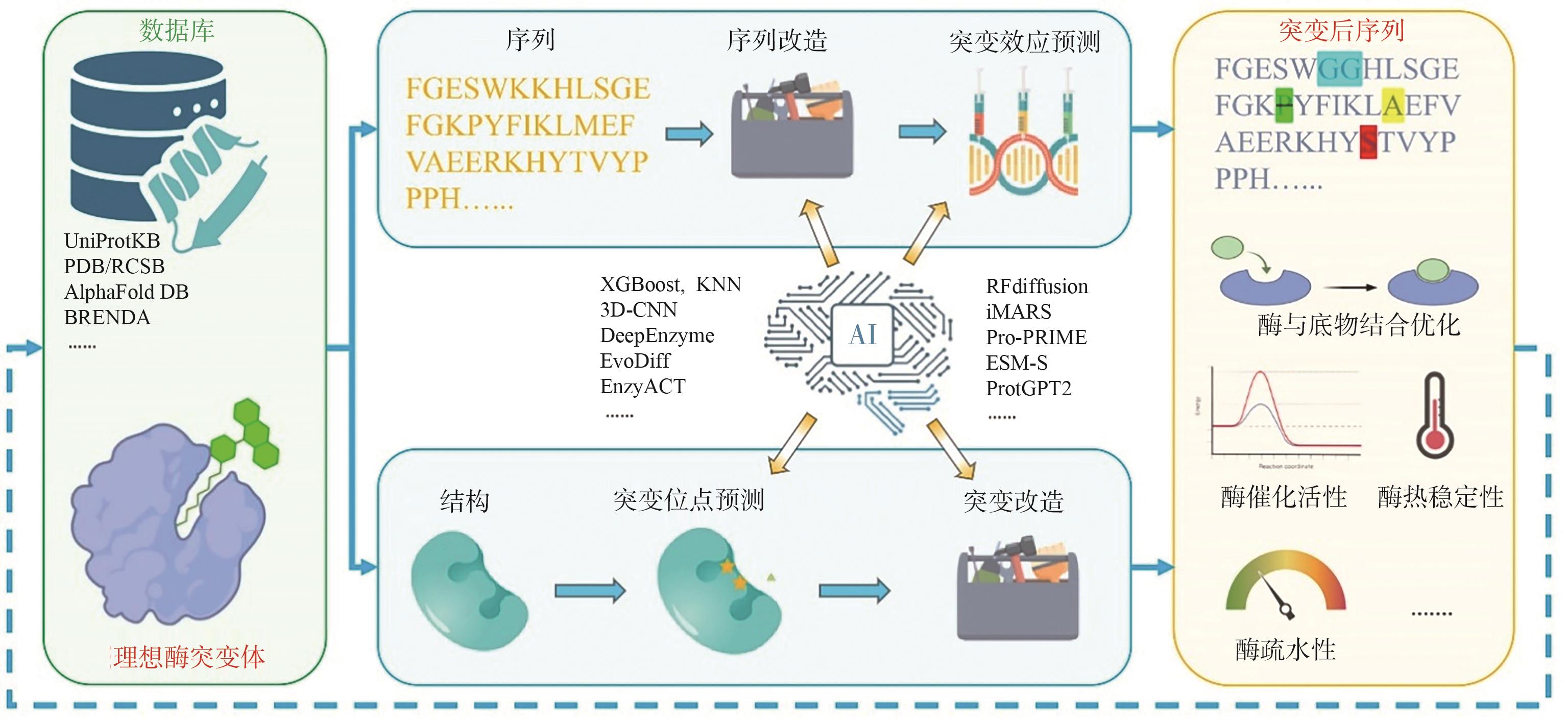
图2 AI驱动的酶改造策略使用已知酶的序列或结构作为输入,进行突变、预测和筛选,最终获得新酶
Fig. 2 AI-driven strategies for enzyme engineeringStarting from known enzyme sequences or structures, this strategy involves mutation, prediction, and screening steps to obtain novel enzyme variants with improved or altered properties
应用类型 Application type | 模型名称 Model name | 模型类型 Model type | 技术特点 Technical features | 发布时间 Publishing time | 参考文献 Reference |
|---|---|---|---|---|---|
| PET水解酶热稳定性、催化活性优化 | MutCompute | 深度学习/机器学习 | 结合机器学习和结构数据来提高PET水解酶的催化性能 | 2022年 | [ |
| 卤代烷烃脱卤素酶和氟化酶性能优化 | MicroPEX-KinMAP | 深度学习/机器学习 | 结合序列和结构生物信息学与微流控技术来发现高效的脱卤酶 | 2022年 | [ |
| 利用机器人自动化与机器学习进行蛋白质定向进化 | BO-EVO | 贝叶斯优化算法 | 聚焦于优化蛋白质的适应性与功能,减轻实验负担 | 2022年 | [ |
| 癌症治疗中的SHP2抑制剂预测 | XGBoost, KNN, 神经网络等 | 深度学习/机器学习 | 通过十倍交叉验证测试多个机器学习模型 | 2023年 | [ |
| 蛋白质-配体相互结合作用优化 | AlphaSpace | 深度学习/机器学习 | 基于AlphaSpace进行靶点预测和功能性优化 | 2023年 | [ |
| 通过多位点组合突变增强果胶裂解酶的热稳定性 | RoseTTAFold | 蛋白质结构预测模型 | 通过迭代设计-测试-学习的方式提升酶的热稳定性 | 2024年 | [ |
| 结合多重突变和蛋白质语言模型优化酶的热稳定性 | Pro-PRIME | 蛋白质语言模型 | 能捕捉到高阶组合突变中的复杂基因互作(表观效应) | 2024年 | [ |
| PET降解水解酶性能重塑与优化 | TurboPETase | 蛋白质语言模型 | TurboPETase重设计使PET降解接近完全,达到200 g/kg的高固体负载 | 2024年 | [ |
| 丝氨酸水解酶局部改造与活性位点优化 | RFdiffusion | 扩散模型(生成模型) | 使用RFdiffusion设计蛋白质活性位点,具有高结构精度 | 2025年 | [ |
| 通过精确的酶结构分析和优化,设计出具有多功能酶活性的复合分子体系 | iMARS | 蛋白质语言模型/生成模型 | iMARS框架不仅适用于生物制造和PET塑料降解,还可以扩展到其他合成生物学和绿色化学领域 | 2025年 | [ |
表2 人工智能在酶改造中的典型方法
Table 2 Representative applications of artificial intelligence in enzyme engineering
应用类型 Application type | 模型名称 Model name | 模型类型 Model type | 技术特点 Technical features | 发布时间 Publishing time | 参考文献 Reference |
|---|---|---|---|---|---|
| PET水解酶热稳定性、催化活性优化 | MutCompute | 深度学习/机器学习 | 结合机器学习和结构数据来提高PET水解酶的催化性能 | 2022年 | [ |
| 卤代烷烃脱卤素酶和氟化酶性能优化 | MicroPEX-KinMAP | 深度学习/机器学习 | 结合序列和结构生物信息学与微流控技术来发现高效的脱卤酶 | 2022年 | [ |
| 利用机器人自动化与机器学习进行蛋白质定向进化 | BO-EVO | 贝叶斯优化算法 | 聚焦于优化蛋白质的适应性与功能,减轻实验负担 | 2022年 | [ |
| 癌症治疗中的SHP2抑制剂预测 | XGBoost, KNN, 神经网络等 | 深度学习/机器学习 | 通过十倍交叉验证测试多个机器学习模型 | 2023年 | [ |
| 蛋白质-配体相互结合作用优化 | AlphaSpace | 深度学习/机器学习 | 基于AlphaSpace进行靶点预测和功能性优化 | 2023年 | [ |
| 通过多位点组合突变增强果胶裂解酶的热稳定性 | RoseTTAFold | 蛋白质结构预测模型 | 通过迭代设计-测试-学习的方式提升酶的热稳定性 | 2024年 | [ |
| 结合多重突变和蛋白质语言模型优化酶的热稳定性 | Pro-PRIME | 蛋白质语言模型 | 能捕捉到高阶组合突变中的复杂基因互作(表观效应) | 2024年 | [ |
| PET降解水解酶性能重塑与优化 | TurboPETase | 蛋白质语言模型 | TurboPETase重设计使PET降解接近完全,达到200 g/kg的高固体负载 | 2024年 | [ |
| 丝氨酸水解酶局部改造与活性位点优化 | RFdiffusion | 扩散模型(生成模型) | 使用RFdiffusion设计蛋白质活性位点,具有高结构精度 | 2025年 | [ |
| 通过精确的酶结构分析和优化,设计出具有多功能酶活性的复合分子体系 | iMARS | 蛋白质语言模型/生成模型 | iMARS框架不仅适用于生物制造和PET塑料降解,还可以扩展到其他合成生物学和绿色化学领域 | 2025年 | [ |

图3 AI驱动的酶设计策略第一种策略为基于序列的方法,其利用深度生成模型学习蛋白质数据集中的共进化模式,并依据数据驱动的原理生成具有潜在功能的序列。第二种策略为基于结构的方法,其通过物理能量函数和空间约束算法,从三维结构限制中推导出稳定的蛋白质构象
Fig. 3 AI-driven strategies for enzyme designSequence-based strategy: Utilizes deep generative models to learn co-evolutionary patterns from protein datasets, and generates novel sequences with potential functional properties in a data-driven manner. Structure-based strategy: Employs physical energy functions and spatial constraint algorithms to derive stable protein conformations based on three-dimensional structural constraints
应用类型 Application type | 模型名称 Model name | 模型类型 Model type | 技术特点 Technical features | 发布时间 Publication time | 参考文献 References |
|---|---|---|---|---|---|
| 通过变分自编码器生成功能性蛋白质变体,应用于酶设计 | MSA-VAE、AR-VAE | 变分自编码器(VAE) | 使用MSA(多序列对齐)和原始序列作为输入,生成新的功能性蛋白变体 | 2021年 | [ |
| 通过生成对抗网络扩展功能性蛋白质序列空间生成新酶 | ProteinGAN | 生成对抗网络(GAN) | 通过自注意力机制学习自然蛋白质序列的进化关系 | 2021年 | [ |
| 蛋白质序列-功能预测与设计,生成性蛋白质设计 | ProT-VAE | 变分自编码器(VAE) | 将VAE与Transformer结合,用于学习序列-功能关系 | 2023年 | [ |
| 使用生成模型设计蛋白质结构与功能 | RFdiffusion | 扩散模型 | 应用于生成具有特定设计目标的功能蛋白,如结合剂设计、酶活性位点支架、对称蛋白体设计 | 2023年 | [ |
| 蛋白质和肽的设计,特别是alpha-螺旋结构的设计 | HelixGAN | 生成对抗网络(GAN) | 通过梯度搜索优化生成的螺旋结构,能够绑定特定靶标或激活细胞受体 | 2023年 | [ |
| 蛋白质和蛋白质复合物的生成,用于蛋白质设计 | Chroma | 扩散模型 | 支持在生成过程中引入多种条件约束(如对称性、形状、语义等) | 2023年 | [ |
| 基于数据的蛋白质设计,生成新的蛋白质序列 | ProtWave-VAE | 变分自编码器(VAE) | 结合VAE和AR模型,在未对齐的序列数据上进行训练和预测 | 2023年 | [ |
| 高亲和力生物活性螺旋肽结合剂设计 | RFdiffusion | 扩散模型 | 能够生成皮摩尔亲和力结合剂用于生物活性肽 | 2024年 | [ |
| 用于从头设计催化新反应的酶 | RFdiffusion2 | 大语言模型 | 可以基于原子级别的活性位点描述设计酶。无需反向旋转生成和预先指定序列位置 | 2025年 | [ |
| 结合大型语言模型(LLMs)和遗传算法(GAs)的新框架,用于加速酶设计,多目标协同优化 | LLM-GA | 大语言模型、遗传算法(GA) | 框架设计高度模块化,可以集成多种性能指标进行酶性能的综合优化 | 2025年 | [ |
表3 人工智能在酶设计中的典型应用
Table 3 Representative applications of artificial intelligence in enzyme design
应用类型 Application type | 模型名称 Model name | 模型类型 Model type | 技术特点 Technical features | 发布时间 Publication time | 参考文献 References |
|---|---|---|---|---|---|
| 通过变分自编码器生成功能性蛋白质变体,应用于酶设计 | MSA-VAE、AR-VAE | 变分自编码器(VAE) | 使用MSA(多序列对齐)和原始序列作为输入,生成新的功能性蛋白变体 | 2021年 | [ |
| 通过生成对抗网络扩展功能性蛋白质序列空间生成新酶 | ProteinGAN | 生成对抗网络(GAN) | 通过自注意力机制学习自然蛋白质序列的进化关系 | 2021年 | [ |
| 蛋白质序列-功能预测与设计,生成性蛋白质设计 | ProT-VAE | 变分自编码器(VAE) | 将VAE与Transformer结合,用于学习序列-功能关系 | 2023年 | [ |
| 使用生成模型设计蛋白质结构与功能 | RFdiffusion | 扩散模型 | 应用于生成具有特定设计目标的功能蛋白,如结合剂设计、酶活性位点支架、对称蛋白体设计 | 2023年 | [ |
| 蛋白质和肽的设计,特别是alpha-螺旋结构的设计 | HelixGAN | 生成对抗网络(GAN) | 通过梯度搜索优化生成的螺旋结构,能够绑定特定靶标或激活细胞受体 | 2023年 | [ |
| 蛋白质和蛋白质复合物的生成,用于蛋白质设计 | Chroma | 扩散模型 | 支持在生成过程中引入多种条件约束(如对称性、形状、语义等) | 2023年 | [ |
| 基于数据的蛋白质设计,生成新的蛋白质序列 | ProtWave-VAE | 变分自编码器(VAE) | 结合VAE和AR模型,在未对齐的序列数据上进行训练和预测 | 2023年 | [ |
| 高亲和力生物活性螺旋肽结合剂设计 | RFdiffusion | 扩散模型 | 能够生成皮摩尔亲和力结合剂用于生物活性肽 | 2024年 | [ |
| 用于从头设计催化新反应的酶 | RFdiffusion2 | 大语言模型 | 可以基于原子级别的活性位点描述设计酶。无需反向旋转生成和预先指定序列位置 | 2025年 | [ |
| 结合大型语言模型(LLMs)和遗传算法(GAs)的新框架,用于加速酶设计,多目标协同优化 | LLM-GA | 大语言模型、遗传算法(GA) | 框架设计高度模块化,可以集成多种性能指标进行酶性能的综合优化 | 2025年 | [ |
| [1] | van Beilen JB, Li Z. Enzyme technology: an overview [J]. Curr Opin Biotechnol, 2002, 13(4): 338-344. |
| [2] | Yang J, Li FZ, Arnold FH. Opportunities and challenges for machine learning-assisted enzyme engineering [J]. ACS Cent Sci, 2024, 10(2): 226-241. |
| [3] | Robinson PK. Enzymes: principles and biotechnological applications [J]. Essays Biochem, 2015, 59: 1-41. |
| [4] | Wiltschi B, Cernava T, Dennig A, et al. Enzymes revolutionize the bioproduction of value-added compounds: From enzyme discovery to special applications [J]. Biotechnol Adv, 2020, 40: 107520. |
| [5] | Victorino da Silva Amatto I, Gonsales da Rosa-Garzon N, Antônio de Oliveira Simões F, et al. Enzyme engineering and its industrial applications [J]. Biotechnol Appl Biochem, 2022, 69(2): 389-409. |
| [6] | Zhou L, Tao CM, Shen XL, et al. Unlocking the potential of enzyme engineering via rational computational design strategies [J]. Biotechnol Adv, 2024, 73: 108376. |
| [7] | Xiong W, Liu B, Shen YJ, et al. Protein engineering design from directed evolution to de novo synthesis [J]. Biochem Eng J, 2021, 174: 108096. |
| [8] | Wang YJ, Xue P, Cao MF, et al. Directed evolution: methodologies and applications [J]. Chem Rev, 2021, 121(20): 12384-12444. |
| [9] | Singh N, Malik S, Gupta A, et al. Revolutionizing enzyme engineering through artificial intelligence and machine learning [J]. Emerg Top Life Sci, 2021, 5(1): 113-125. |
| [10] | Mao SC, Jiang JW, Xiong K, et al. Enzyme engineering: performance optimization, novel sources, and applications in the food industry [J]. Foods, 2024, 13(23): 3846. |
| [11] | Xu KJ, Fu HR, Chen QM, et al. Engineering thermostability of industrial enzymes for enhanced application performance [J]. Int J Biol Macromol, 2025, 291: 139067. |
| [12] | Srivastava N, Khare SK. Advances in microbial alkaline proteases: addressing industrial bottlenecks through genetic and enzyme engineering [J]. Appl Biochem Biotechnol, 2025, 197(8): 4861-4896. |
| [13] | Sikander R, Wang YP, Ghulam A, et al. Identification of enzymes-specific protein domain based on DDE, and convolutional neural network [J]. Front Genet, 2021, 12: 759384. |
| [14] | Dae J, Bae K, Kim Y, et al. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering [J]. Curr Opin Biotechnol, 2022, 73: 101-107. |
| [15] | Wang YH, Han SX, Wang Y, et al. Artificial intelligence technology assists enzyme prediction and rational design [J]. J Agric Food Chem, 2025, 73(12): 7065-7073. |
| [16] | Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021, 596(7873): 583-589. |
| [17] | Watson JL, Juergens D, Bennett NR, et al. De novo design of protein structure and function with RFdiffusion [J]. Nature, 2023, 620(7976): 1089-1100. |
| [18] | Sun MGF, Seo MH, Nim S, et al. Protein engineering by highly parallel screening of computationally designed variants [J]. Sci Adv, 2016, 2(7): e1600692. |
| [19] | Siedhoff NE, Schwaneberg U, Davari MD. Machine learning-assisted enzyme engineering [J]. Meth Enzymol, 2020, 643: 281-315. |
| [20] | Strokach A, Kim PM. Deep generative modeling for protein design [J]. Curr Opin Struct Biol, 2022, 72: 226-236. |
| [21] | Corso G, Stark H, Jegelka S, et al. Graph neural networks [J]. Nat Rev Meth Primers, 2024, 4: 17. |
| [22] | Hsu C, Fannjiang C, Listgarten J. Generative models for protein structures and sequences [J]. Nat Biotechnol, 2024, 42(2): 196-199. |
| [23] | Fram B, Su Y, Truebridge I, et al. Simultaneous enhancement of multiple functional properties using evolution-informed protein design [J]. Nat Commun, 2024, 15: 5141. |
| [24] | Chen Z, Liu YG, Wang YG, et al. Validation of an LLM-based multi-agent framework for protein engineering in dry lab and wet lab [C]//2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). December 3-6, 2024, Lisbon, Portugal. Piscataway, NJ: IEEE, 2024: 5364-5370. |
| [25] | Wildey MJ, Haunso A, Tudor M, et al. High-throughput screening [M]//Platform Technologies in Drug Discovery and Validation. Amsterdam: Elsevier, 2017: 149-195. |
| [26] | Fowler DM, Fields S. Deep mutational scanning: a new style of protein science [J]. Nat Meth, 2014, 11(8): 801-807. |
| [27] | Lee SO, Fried SD. An error prone PCR method for small amplicons [J]. Anal Biochem, 2021, 628: 114266. |
| [28] | Giessel A, Dousis A, Ravichandran K, et al. Therapeutic enzyme engineering using a generative neural network [J]. Sci Rep, 2022, 12: 1536. |
| [29] | Li FR, Yuan L, Lu HZ, et al. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction [J]. Nat Catal, 2022, 5(8): 662-672. |
| [30] | Varadi M, Anyango S, Deshpande M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models [J]. Nucleic Acids Res, 2022, 50(D1): D439-D444. |
| [31] | Boutet E, Lieberherr D, Tognolli M, et al. UniProtKB/Swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view [M]//Plant Bioinformatics. New York, NY: Springer New York, 2016: 23-54. |
| [32] | Bittrich S, Bhikadiya C, Bi CX, et al. RCSB protein data bank: efficient searching and simultaneous access to one million computed structure models alongside the PDB structures enabled by architectural advances [J]. J Mol Biol, 2023, 435(14): 167994. |
| [33] | Nikam R, Kulandaisamy A, Harini K, et al. ProThermDB: thermodynamic database for proteins and mutants revisited after 15 years [J]. Nucleic Acids Res, 2021, 49(D1): D420-D424. |
| [34] | Stourac J, Dubrava J, Musil M, et al. FireProtDB: database of manually curated protein stability data [J]. Nucleic Acids Res, 2021, 49(D1): D319-D324. |
| [35] | Velecký J, Hamsikova M, Stourac J, et al. SoluProtMutDB: a manually curated database of protein solubility changes upon mutations [J]. Comput Struct Biotechnol J, 2022, 20: 6339-6347. |
| [36] | Wang CY, Chang PM, Ary ML, et al. ProtaBank: a repository for protein design and engineering data [J]. Protein Sci, 2018, 27(6): 1113-1124. |
| [37] | Hunter S, Apweiler R, Attwood TK, et al. InterPro: the integrative protein signature database [J]. Nucleic Acids Res, 2009, 37(): D211-D215. |
| [38] | Schomburg I, Chang A, Hofmann O, et al. BRENDA: a resource for enzyme data and metabolic information [J]. Trends Biochem Sci, 2002, 27(1): 54-56. |
| [39] | Sankaranarayanan K, Jensen KF. Computer-assisted multistep chemoenzymatic retrosynthesis using a chemical synthesis planner [J]. Chem Sci, 2023, 14(23): 6467-6475. |
| [40] | Zhou ZY, Zhang L, Yu YX, et al. Enhancing efficiency of protein language models with minimal wet-lab data through few-shot learning [J]. Nat Commun, 2024, 15: 5566. |
| [41] | Zhang L, Luo K, Zhou ZY, et al. A deep retrieval-enhanced meta-learning framework for enzyme optimum pH prediction [J]. J Chem Inf Model, 2025, 65(7): 3761-3770. |
| [42] | Patsch D, Buller R. Improving enzyme fitness with machine learning [J]. Chimia, 2023, 77(3): 116. |
| [43] | Wei SZ, Chen ZY, Arumugasamy SK, et al. Data augmentation and machine learning techniques for control strategy development in bio-polymerization process [J]. Environ Sci Ecotechnol, 2022, 11: 100172. |
| [44] | Xie XZ, Valiente PA, Kim PM. HelixGAN a deep-learning methodology for conditional de novo design of α-helix structures [J]. Bioinformatics, 2023, 39(1): btad036. |
| [45] | 徐沛,汪卫华,宁洪伟, 等. 人工智能辅助的酶分子改造应用进展[J]. 生物工程学报, 2024, 40(6): 1728-1741. |
| Xu P, Wang W, Ning H, et al. Progress in the application of artificial intelligence-assisted molecular modification of enzymes [J]. Chinese Journal of Biotechnology, 2024, 40(6): 1728-1741. | |
| [46] | Biswas S, Khimulya G, Alley EC, et al. Low-N protein engineering with data-efficient deep learning [J]. Nat Meth, 2021, 18(4): 389-396. |
| [47] | Shroff R, Cole AW, Diaz DJ, et al. Discovery of novel gain-of-function mutations guided by structure-based deep learning [J]. ACS Synth Biol, 2020, 9(11): 2927-2935. |
| [48] | Brandes N, Ofer D, Peleg Y, et al. ProteinBERT: a universal deep-learning model of protein sequence and function [J]. Bioinformatics, 2022, 38(8): 2102-2110. |
| [49] | Alexander Rives JM. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences [J]. Proc Natl Acad Sci U S A, 2021, 118(15): 1-12. |
| [50] | Rao RM, Liu J, Verkuil R, et al. MSA transformer [C]//Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 2021. |
| [51] | Language models enable zero-shot prediction of the effects of mutations on protein function [C]//Proceedings of the 35th International Conference on Neural Information Processing Systems. New York: ACM, 2021: 29287-29303. |
| [52] | Lin ZM, Akin H, Rao R, et al. Evolutionary-scale prediction of atomic-level protein structure with a language model [J]. Science, 2023, 379(6637): 1123-1130. |
| [53] | Lu HY, Diaz DJ, Czarnecki NJ, et al. Machine learning-aided engineering of hydrolases for PET depolymerization [J]. Nature, 2022, 604(7907): 662-667. |
| [54] | Vasina M, Vanacek P, Hon J, et al. Advanced database mining of efficient haloalkane dehalogenases by sequence and structure bioinformatics and microfluidics [J]. Chem Catal, 2022, 2(10): 2704-2725. |
| [55] | Hu RY, Fu LH, Chen YC, et al. Protein engineering via Bayesian optimization-guided evolutionary algorithm and robotic experiments [J]. Brief Bioinform, 2023, 24(1): bbac570. |
| [56] | Adhikari N, Ayyannan SR. Development and validation of machine learning models for the prediction of SH-2 containing protein tyrosine phosphatase 2 inhibitors [J]. Mol Divers, 2024, 28(4): 1889-1905. |
| [57] | Xia S, Chen E, Zhang YK. Integrated molecular modeling and machine learning for drug design [J]. J Chem Theory Comput, 2023, 19(21): 7478-7495. |
| [58] | Zhang ZH, Li ZX, Yang ML, et al. Machine learning-guided multi-site combinatorial mutagenesis enhances the thermostability of pectin lyase [J]. Int J Biol Macromol, 2024, 277: 134530. |
| [59] | Bian JH, Tan P, Nie T, et al. Optimizing enzyme thermostability by combining multiple mutations using protein language model [J]. mLife, 2024, 3(4): 492-504. |
| [60] | Cui YL, Chen YC, Sun JY, et al. Computational redesign of a hydrolase for nearly complete PET depolymerization at industrially relevant high-solids loading [J]. Nat Commun, 2024, 15: 1417. |
| [61] | Lauko A, Pellock SJ, Sumida KH, et al. Computational design of serine hydrolases [J]. Science, 2025, 388(6744): eadu2454. |
| [62] | Wang JW, Ouyang XY, Meng SY, et al. Rational multienzyme architecture design with iMARS [J]. Cell, 2025, 188(5): 1349-1362.e17. |
| [63] | Lutz ID, Wang SZ, Norn C, et al. Top-down design of protein architectures with reinforcement learning [J]. Science, 2023, 380(6642): 266-273. |
| [64] | Offline reinforcement learning as one big sequence modeling problem [C]//Proceedings of the 35th International Conference on Neural Information Processing Systems. New York: ACM, 2021: 1273-1286. |
| [65] | Wang CR, Chen Y, Zhang Y, et al. A reinforcement learning approach for protein-ligand binding pose prediction [J]. BMC Bioinform, 2022, 23(1): 368. |
| [66] | Jha K, Saha S, Singh H. Prediction of protein-protein interaction using graph neural networks [J]. Sci Rep, 2022, 12: 8360. |
| [67] | Khan S, Noor S, Awan HH, et al. Deep-ProBind: binding protein prediction with transformer-based deep learning model [J]. BMC Bioinform, 2025, 26(1): 88. |
| [68] | Wang T, Xiang GM, He SW, et al. DeepEnzyme: a robust deep learning model for improved enzyme turnover number prediction by utilizing features of protein 3D-structures [J]. Brief Bioinform, 2024, 25(5): bbae409. |
| [69] | Li G, Zhang N, Dai XW, et al. EnzyACT: a novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity [J]. J Chem Inf Model, 2024, 64(15): 5912-5921. |
| [70] | Jiang Y, Ran XC, Yang ZJ. Data-driven enzyme engineering to identify function-enhancing enzymes [J]. Protein Eng Des Sel, 2023, 36: gzac009. |
| [71] | Wang XR, Yin XD, Jiang DJ, et al. Multi-modal deep learning enables efficient and accurate annotation of enzymatic active sites [J]. Nat Commun, 2024, 15: 7348. |
| [72] | Abdine H, Chatzianastasis M, Bouyioukos C, et al. Prot2Text: multimodal protein’s function generation with GNNs and transformers [J]. Proc AAAI Conf Artif Intell, 2024, 38(10): 10757-10765. |
| [73] | Schlichtkrull M, Kipf TN, Bloem P, et al. Modeling relational data with graph convolutional networks [C]//The Semantic Web. New York: ACM, 2018: 593-607. |
| [74] | Ahern W, Yim J, Tischer D, et al. Atom level enzyme active site scaffolding using RFdiffusion2[J]. bioRxiv, 2025: 2025.04. 09. 648075. |
| [75] | Kim D, Noh MH, Park M, et al. Enzyme activity engineering based on sequence co-evolution analysis [J]. Metab Eng, 2022, 74: 49-60. |
| [76] | Hawkins-Hooker A, Depardieu F, Baur S, et al. Generating functional protein variants with variational autoencoders [J]. PLoS Comput Biol, 2021, 17(2): e1008736. |
| [77] | Repecka D, Jauniskis V, Karpus L, et al. Expanding functional protein sequence spaces using generative adversarial networks [J]. Nat Mach Intell, 2021, 3(4): 324-333. |
| [78] | Sevgen E, Moller J, Lange A, et al. ProT-VAE: protein transformer variational autoencoder for functional protein design [J]. bioRxiv, 2023. DOI:10.1101/2023.01.23.525232 |
| [79] | Ingraham JB, Baranov M, Costello Z, et al. Illuminating protein space with a programmable generative model [J]. Nature, 2023, 623(7989): 1070-1078. |
| [80] | Praljak N, Lian XR, Ranganathan R, et al. ProtWave-VAE: integrating autoregressive sampling with latent-based inference for data-driven protein design [J]. ACS Synth Biol, 2023, 12(12): 3544-3561. |
| [81] | Vázquez Torres S, Leung PJY, Venkatesh P, et al. De novo design of high-affinity binders of bioactive helical peptides [J]. Nature, 2024, 626(7998): 435-442. |
| [82] | Nana Teukam YG, Zipoli F, Laino T, et al. Integrating genetic algorithms and language models for enhanced enzyme design [J]. Brief Bioinform, 2024, 26(1): bbae675. |
| [83] | Boob AG, Tan SI, Zaidi A, et al. Design of diverse, functional mitochondrial targeting sequences across eukaryotic organisms using variational autoencoder [J]. Nat Commun, 2025, 16: 4151. |
| [84] | Ingraham J, Garg VK, Barzilay R, et al. Generative models for graph-based protein design [J]. 2019: 15820-15831. |
| [85] | Dauparas J, Anishchenko I, Bennett N, et al. Robust deep learning-based protein sequence design using ProteinMPNN [J]. Science, 2022, 378(6615): 49-56. |
| [86] | Yang JY, Anishchenko I, Park H, et al. Improved protein structure prediction using predicted interresidue orientations [J]. Proc Natl Acad Sci U S A, 2020, 117(3): 1496-1503. |
| [87] | Hansen AL, Theisen FF, Crehuet R, et al. Carving out a glycoside hydrolase active site for incorporation into a new protein scaffold using deep network hallucination [J]. ACS Synth Biol, 2024, 13(3): 862-875. |
| [88] | Baek M, DiMaio F, Anishchenko I, et al. Accurate prediction of protein structures and interactions using a three-track neural network [J]. Science, 2021, 373(6557): 871-876. |
| [89] | Joho Y, Royan S, Caputo AT, et al. Enhancing PET degrading enzymes: a combinatory approach [J]. ChemBioChem, 2024, 25(10): e202400084. |
| [90] | Xi Y, Ye LD, Yu HW. Enhanced thermal and alkaline stability of L-lysine decarboxylase CadA by combining directed evolution and computation-guided virtual screening [J]. Bioresour Bioprocess, 2022, 9(1): 24. |
| [91] | Scherer M, Fleishman SJ, Jones PR, et al. Computational enzyme engineering pipelines for optimized production of renewable chemicals [J]. Front Bioeng Biotechnol, 2021, 9: 673005. |
| [92] | Srinivas N, Krause A, Kakade S M, et al. Gaussian process optimization in the bandit setting: No regret and experimental design [J]. Cornell University Library, 2010. DOI: 10.48550/arxiv.0912.3995 |
| [93] | Fenoy E, Edera AA, Stegmayer G. Transfer learning in proteins: evaluating novel protein learned representations for bioinformatics tasks [J]. Brief Bioinform, 2022, 23(4): bbac232. |
| [94] | Pachter R, Wang ZQ. Adaptive simulated annealing and its application to protein folding [M]//Encyclopedia of Optimization. Cham: Springer Nature Switzerland, 2024: 1-6. |
| [95] | Narayanan H, Dingfelder F, Butté A, et al. Machine learning for biologics: opportunities for protein engineering, developability, and formulation [J]. Trends Pharmacol Sci, 2021, 42(3): 151-165. |
| [96] | Ge FG, Gao YH, Jiang YJ, et al. Design and performance analysis of multi-enzyme activity-doped nanozymes assisted by machine learning [J]. Colloids Surf B Biointerfaces, 2025, 248: 114468. |
| [97] | Ding K, Chin M, Zhao YL, et al. Machine learning-guided co-optimization of fitness and diversity facilitates combinatorial library design in enzyme engineering [J]. Nat Commun, 2024, 15: 6392. |
| [98] | Zimmerman L, Alon N, Levin I, et al. Context-dependent design of induced-fit enzymes using deep learning generates well-expressed, thermally stable and active enzymes [J]. Proc Natl Acad Sci U S A, 2024, 121(11): e2313809121. |
| [99] | Xu YY, Zhao XJ, Song XZ, et al. Boosting protein language models with negative sample mining [M]//Machine Learning and Knowledge Discovery in Databases. Applied Data Science Track. Cham: Springer Nature Switzerland, 2024: 199-214. |
| [100] | Chatterjee A, Ravandi B, Haddadi P, et al. Topology-driven negative sampling enhances generalizability in protein-protein interaction prediction [J]. Bioinformatics, 2025, 41(5): btaf148. |
| [101] | Niazi SK. Protein catalysis through structural dynamics: a comprehensive analysis of energy conversion in enzymatic systems and its computational limitations [J]. Pharmaceuticals, 2025, 18(7): 951. |
| [102] | Qin YM, Chen ZH, Peng Y, et al. Deep learning methods for protein structure prediction [J]. MedComm, 2024, 3(3): e96. |
| [103] | Xie WJ, Warshel A. Harnessing generative AI to decode enzyme catalysis and evolution for enhanced engineering [J]. bioRxiv, 2023: 2023.10.10.561808. |
| [1] | 王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53. |
| [2] | 陈强, 于璎霏, 张颖, 张冲. 茉莉酸甲酯对薄皮甜瓜‘绿宝石’采后冷害的调控[J]. 生物技术通报, 2025, 41(9): 105-114. |
| [3] | 于文杰, 范斯然, 高文丽, 邢宇, 秦岭. 板栗核黄素合成通路关键基因鉴定及功能验证[J]. 生物技术通报, 2025, 41(9): 232-241. |
| [4] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [5] | 廉少杰, 唐胜硕, 康传利, 刘磊, 郑德强, 杜帅, 汤丽伟, 张美霞, 刘蔷. 高产银耳多糖酶菌株的分离、鉴定、发酵条件优化及其酶的特性分析[J]. 生物技术通报, 2025, 41(9): 302-313. |
| [6] | 张永艳, 郭思健, 李晶, 郝思怡, 李瑞得, 刘嘉鹏, 程春振. 蓝莓花青素相关VcGSTF19基因的克隆及功能研究[J]. 生物技术通报, 2025, 41(9): 139-146. |
| [7] | 李亚涛, 张志鹏, 赵梦瑶, 吕镇, 甘恬, 魏浩, 吴书凤, 马玉超. 根瘤菌Bd1的全基因组分析及TetR3对细胞生长和结瘤的负调控功能[J]. 生物技术通报, 2025, 41(9): 289-301. |
| [8] | 郑乾明, 晏霜, 解璞, 王红林. 红肉火龙果细胞壁转化酶基因SmCWIN6的表达和酶活性鉴定[J]. 生物技术通报, 2025, 41(8): 267-275. |
| [9] | 蔡如凤, 杨宇轩, 于基正, 李佳楠. 人工智能重塑蛋白质工程:从结构解析到合成生物学的算法革命[J]. 生物技术通报, 2025, 41(8): 1-10. |
| [10] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [11] | 刁辰洋, 崔有志, 李炳志. 靶向诱变介导的微生物进化技术研究进展[J]. 生物技术通报, 2025, 41(8): 11-21. |
| [12] | 李加仪, 李尽益, 白雪, 柏映国, 刘波, 张志伟. 稀有密码子串联介导的HemB表达弱化提升5-氨基乙酰丙酸的含量[J]. 生物技术通报, 2025, 41(8): 74-81. |
| [13] | 王辉, 范灵熙, 孙纪录, 王苑, 伍宁丰, 田健, 关菲菲. 基于蛋白智能模型提升溶菌酶RPL187的热稳定性[J]. 生物技术通报, 2025, 41(7): 336-346. |
| [14] | 王从欢, 伍国强, 魏明. 植物CBL调控逆境胁迫响应的作用机制[J]. 生物技术通报, 2025, 41(7): 1-16. |
| [15] | 黄旭升, 周雅莉, 柴旭东, 闻婧, 王计平, 贾小云, 李润植. 紫苏质体型PfLPAT1B基因的克隆及其在油脂合成中的功能分析[J]. 生物技术通报, 2025, 41(7): 226-236. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||