








生物技术通报 ›› 2022, Vol. 38 ›› Issue (1): 141-149.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0418
收稿日期:2021-03-31
出版日期:2022-01-26
发布日期:2022-02-22
作者简介:孙靖雅,硕士研究生,研究方向:资源与环境微生物;E-mail: Received:2021-03-31
Published:2022-01-26
Online:2022-02-22
摘要:
长期在砷污染环境生存的细菌逐渐演化出抗砷机制,旨在评价分离自湖南铅锌矿区污染土壤的Pseudomonas sp. Tw224抗砷能力和抗重金属谱;揭示该菌的抗砷基因/基因簇的功能。在砷或重金属存在条件下测量细菌的生长情况,对该菌的抗砷能力和抗重金属谱进行评价;通过基因组框架图分析该菌的抗砷基因/基因簇;采用代谢工程法构建基因簇缺失突变株揭示基因簇的功能。结果显示,砷延长了该菌的延迟期,降低了该菌的最高菌浓,该菌能够抗As5+和As3+的最高浓度分别为600 mmol/L和20 mmol/L,并且对铜、镍、锌、镉、铬、银和汞均具有不同程度的抗性。该菌基因组中共包含了20个与抗砷相关的基因(1个arsH,4个arsC,arsB和ACR(3)各2个,11个arsR),其中arsH-arsC2-arsB-arsR(ars1)和arsR-arsB-arsC2(ars2)分别以基因簇的形式存在,长度分别为2.8 kb和2.2 kb。采用同源重组双交换法成功构建了ars1、ars2单缺失和ars1-ars2双缺失突变株Pseudomonas sp. QSA1、Pseudomonas sp. QSA2和Pseudomonas sp. QSARS。在无砷条件下3株突变株的生长均未受到影响,砷对QSA1和QSA2的最低抑菌浓度分别降低为野生菌Tw224的4%和80%;反应48 h,这两株突变株菌的砷还原能力分别约为野生菌的9.8%和53.8%。双基因簇突变株QSARS几乎完全失去了抗砷和砷的还原能力。Tw224具有很强的抗砷能力和广谱的抗重金属能力;基因组中存在大量抗重金属相关基因,其中有两个抗砷相关基因簇,ars1起主要的抗砷作用,ars2起辅助功能。
孙靖雅, 马玉超. 假单胞菌Tw224抗砷基因簇的功能研究[J]. 生物技术通报, 2022, 38(1): 141-149.
SUN Jing-ya, MA Yu-chao. Functions of Arsenic-resistant Gene Cluster in Pseudomonas sp. Tw224[J]. Biotechnology Bulletin, 2022, 38(1): 141-149.
| 质粒和菌株Plasmids and strains | 特性Characteristic | 来源Source | |
|---|---|---|---|
| 质粒 Plasmid | pK18mobsacB | Suicide plasmid vector,KanR | Stored in lab |
| pSET152 | Integrativeplasmid vector,Apr | Stored in lab | |
| pA18mobsacB | Apr | This study | |
| pA18QCA1 | pA18 with a1 up/a1 down,Apr | This study | |
| pA18QCA2 | pA18 with a2 up/a2 down,Apr | This study | |
| 菌株 Strain | Pseudomonas sp. Tw224 | KanR、AmpR | From Hunan,stored in lab |
| E. coli S17-1 | KanR | This study | |
| E. coli S17-1-QCA1 | E. coli S17-1 with pA18QCA1 | This study | |
| E. coli S17-1-QCA2 | E. coli S17-1 with pA18QCA2 | This study | |
| Pseudomonas sp. QSA1 | Tw224 ∆ars1 | This study | |
| Pseudomonas sp. QSA2 | Tw224 ∆ars2 | This study | |
| Pseudomonas sp. QSARS | Tw224 ∆ars1 ars2 | This study | |
表1 实验中使用的菌株和质粒
Table 1 Plasmids and strains in this study
| 质粒和菌株Plasmids and strains | 特性Characteristic | 来源Source | |
|---|---|---|---|
| 质粒 Plasmid | pK18mobsacB | Suicide plasmid vector,KanR | Stored in lab |
| pSET152 | Integrativeplasmid vector,Apr | Stored in lab | |
| pA18mobsacB | Apr | This study | |
| pA18QCA1 | pA18 with a1 up/a1 down,Apr | This study | |
| pA18QCA2 | pA18 with a2 up/a2 down,Apr | This study | |
| 菌株 Strain | Pseudomonas sp. Tw224 | KanR、AmpR | From Hunan,stored in lab |
| E. coli S17-1 | KanR | This study | |
| E. coli S17-1-QCA1 | E. coli S17-1 with pA18QCA1 | This study | |
| E. coli S17-1-QCA2 | E. coli S17-1 with pA18QCA2 | This study | |
| Pseudomonas sp. QSA1 | Tw224 ∆ars1 | This study | |
| Pseudomonas sp. QSA2 | Tw224 ∆ars2 | This study | |
| Pseudomonas sp. QSARS | Tw224 ∆ars1 ars2 | This study | |
| 引物名称 Primer | 序列 Sequence(5'-3') |
|---|---|
| Apr UP | cgcagctgtgctcgacgtGAAGATCCTTTGATCTTTTC |
| Apr DOWN | ttcggcaagcaggcatcgccatgATCAGCCGTCCAAATGC |
| MQa1U S | attcgagctcggtacccgggAGGACCAGGCTGAGGACA |
| MQa1U A | TATTTGGTAGACCGCTATTCG |
| MQa1D S | gaatagcggtctaccaaataCCCAAGCACTTGAAGAATGT |
| MQa1D A | taaaacgacggccagtgccaCTGCCGAGCAAAGCGTAT |
| NEW2U S | attcgagctcggtacccgggCAGGGACAAGGGAATGACG |
| NEW2U A | TTGACGACGGTCCACCAG |
| MQa2D S | ctggtggaccgtcgtcaaAGGCGCTGATTCTGTGGG |
| MQa2D A | taaaacgacggccagtgccaTCAATGAGGCTGCGGATG |
| YZ UP | CCCCAGGCTTTACACTTTATG |
| YZ DOWN | TGTGCTGCAAGGCGATTA |
| YZA1Y S | ACCGATGTGAGCGAAGCC |
| YZA1Y A | CCTCCTGGTCGTTACCCTGT |
| YZA2Y S | CAATCAGTTGTGGCGGTTTC |
| YZA2Y A | CATGCTCAGCTCCTTTCGA |
表2 实验中使用的引物
Table 2 Primers in this study
| 引物名称 Primer | 序列 Sequence(5'-3') |
|---|---|
| Apr UP | cgcagctgtgctcgacgtGAAGATCCTTTGATCTTTTC |
| Apr DOWN | ttcggcaagcaggcatcgccatgATCAGCCGTCCAAATGC |
| MQa1U S | attcgagctcggtacccgggAGGACCAGGCTGAGGACA |
| MQa1U A | TATTTGGTAGACCGCTATTCG |
| MQa1D S | gaatagcggtctaccaaataCCCAAGCACTTGAAGAATGT |
| MQa1D A | taaaacgacggccagtgccaCTGCCGAGCAAAGCGTAT |
| NEW2U S | attcgagctcggtacccgggCAGGGACAAGGGAATGACG |
| NEW2U A | TTGACGACGGTCCACCAG |
| MQa2D S | ctggtggaccgtcgtcaaAGGCGCTGATTCTGTGGG |
| MQa2D A | taaaacgacggccagtgccaTCAATGAGGCTGCGGATG |
| YZ UP | CCCCAGGCTTTACACTTTATG |
| YZ DOWN | TGTGCTGCAAGGCGATTA |
| YZA1Y S | ACCGATGTGAGCGAAGCC |
| YZA1Y A | CCTCCTGGTCGTTACCCTGT |
| YZA2Y S | CAATCAGTTGTGGCGGTTTC |
| YZA2Y A | CATGCTCAGCTCCTTTCGA |
| 菌株Strain | 最低抑菌浓度MIC /(mmol·L-1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| As5+ | As3+ | Cr3+ | Ag3+ | Cd2+ | Co2+ | Ni2+ | Hg2+ | Zn2+ | Cu2+ | ||
| Tw224 | 600 | 20 | 5 | 3 | 2 | 3 | 12 | 3 | 12 | 20 | |
表3 Pseudomonas sp. Tw224对8种重金属的MIC值
Table 3 MIC to 8 heavy metals of Pseudomonas sp. Tw224
| 菌株Strain | 最低抑菌浓度MIC /(mmol·L-1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| As5+ | As3+ | Cr3+ | Ag3+ | Cd2+ | Co2+ | Ni2+ | Hg2+ | Zn2+ | Cu2+ | ||
| Tw224 | 600 | 20 | 5 | 3 | 2 | 3 | 12 | 3 | 12 | 20 | |
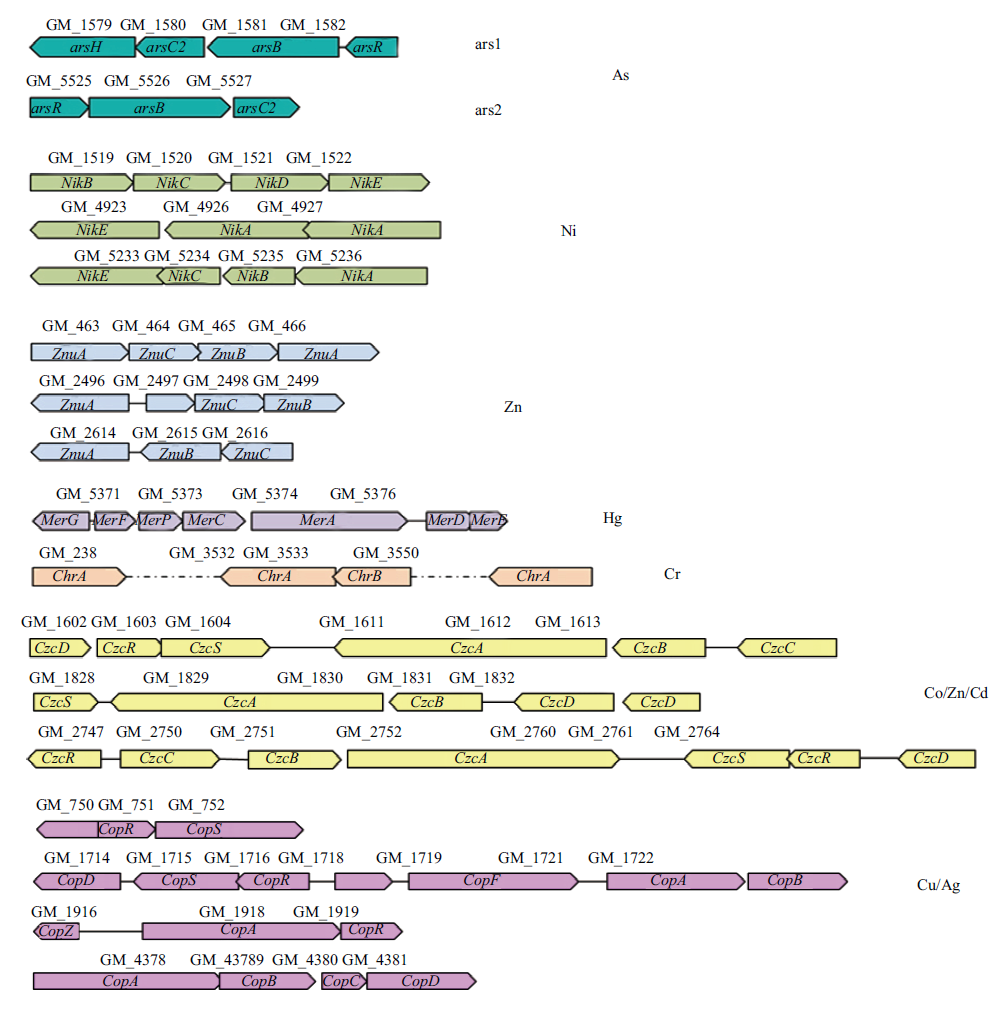
图2 Pseudomonas sp. Tw224中与砷与其他重金属抗性相关的基因和基因簇
Fig. 2 Genes and gene clusters related to resistances to arsenic and other heavy metals in Pseudomonas sp. Tw224
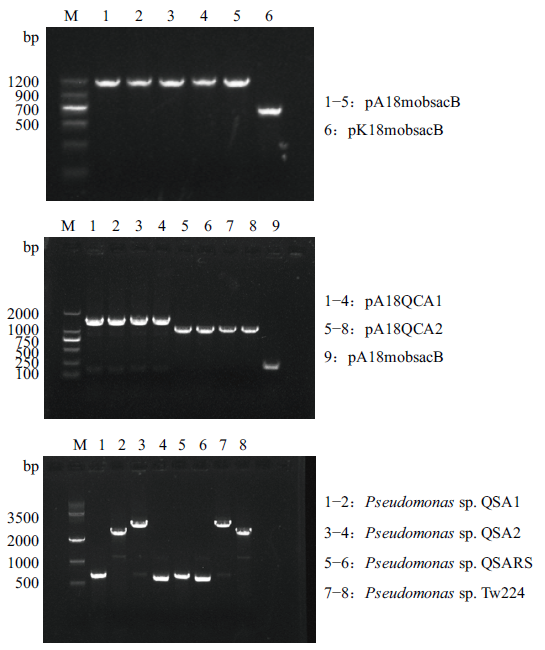
图3 各重组质粒及突变株的验证 :pA18mobsacB的验证;B:pA18QCA1/pA18QCA2的验证;C:Pseudomonas sp. QSA1/Pseudomonas sp. QSA2/Pseudomonas sp. QSARS的验证
Fig. 3 Verification of recombinant plasmids and mutants
| [1] |
Mandal BK, Suzuki KT. Arsenic round the world:a review[J]. Talanta, 2002, 58(1):201-235.
pmid: 18968746 |
| [2] |
Zhu YG, Yoshinaga M, et al. Earth abides arsenic biotransformations[J]. Annu Rev Earth Planet Sci, 2014, 42(1):443-467.
doi: 10.1146/earth.2014.42.issue-1 URL |
| [3] |
Mestrot A, Feldmann J, Krupp EM, et al. Field fluxes and speciation of arsines emanating from soils[J]. Environ Sci Technol, 2011, 45(5):1798-1804.
doi: 10.1021/es103463d pmid: 21284382 |
| [4] |
Cavalca L, Corsini A, Zaccheo P, et al. Microbial transformations of arsenic:perspectives for biological removal of arsenic from water[J]. Future Microbiol, 2013, 8(6):753-768.
doi: 10.2217/fmb.13.38 pmid: 23586329 |
| [5] |
Sousa T, Branco R, Piedade AP, et al. Hyper accumulation of arsenic in mutants of Ochrobactrum tritici silenced for arsenite efflux pumps[J]. PLoS One, 2015, 10(7):e0131317.
doi: 10.1371/journal.pone.0131317 URL |
| [6] |
Huang H, Jia Y, Sun GX, et al. Arsenic speciation and volatilization from flooded paddy soils amended with different organic matters[J]. Environ Sci Technol, 2012, 46(4):2163-2168.
doi: 10.1021/es203635s pmid: 22295880 |
| [7] |
Li J, Pawitwar SS, Rosen BP. The organoarsenical biocycle and the primordial antibiotic methylarsenite[J]. Metallomics, 2016, 8(10):1047-1055.
doi: 10.1039/C6MT00168H URL |
| [8] |
Cai L, Liu G, Rensing C, et al. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils[J]. BMC Microbiol, 2009, 9:4.
doi: 10.1186/1471-2180-9-4 URL |
| [9] | Yang HC, Fu HL, Lin YF, et al. Pathways of arsenic uptake and efflux[J]. Curr Top Membr, 2012, 69:325-358. |
| [10] | 陈倩, 苏建强, 等. 微生物砷还原机制的研究进展[J]. 生态毒理学报, 2011, 6(3):225-233. |
| Chen Q, Su JQ, et al. Advances in mechanisms of microbial arsenate reduction[J]. Asian J Ecotoxicol, 2011, 6(3):225-233. | |
| [11] |
Páez-Espino AD, Nikel PI, Chavarría M, et al. ArsH protects Pseudomonas putida from oxidative damage caused by exposure to arsenic[J]. Environ Microbiol, 2020, 22(6):2230-2242.
doi: 10.1111/1462-2920.14991 pmid: 32202357 |
| [12] |
Wu D, Zhang Z, Gao Q, et al. Isolation and characterization of aerobic, culturable, arsenic-tolerant bacteria from lead-zinc mine tailing in Southern China[J]. World J Microbiol Biotechnol, 2018, 34(12):177.
doi: 10.1007/s11274-018-2557-x URL |
| [13] | 吕常江, 赵春贵, 杨素萍, 等. 紫色非硫细菌的砷代谢机制[J]. 微生物学报, 2012, 52(12):1497-1507. |
| Lv CJ, Zhao CG, Yang SP, et al. Arsenic metabolism in purple nonsulfur bacteria[J]. Acta Microbiol Sin, 2012, 52(12):1497-1507. | |
| [14] |
Chong TM, Yin WF, et al. Comprehensive genomic and phenotypic metal resistance profile of Pseudomonas putida strain S13. 1. 2 isolated from a vineyard soil[J]. AMB Express, 2016, 6(1):95.
doi: 10.1186/s13568-016-0269-x pmid: 27730570 |
| [15] |
Páez-Espino AD, Durante-Rodríguez G, de Lorenzo V. Functional coexistence of twin arsenic resistance systems in Pseudomonas putida KT2440[J]. Environ Microbiol, 2015, 17(1):229-238.
doi: 10.1111/1462-2920.12464 pmid: 24673935 |
| [16] |
Koechler S, Arsène-Ploetze F, Brochier-Armanet C, et al. Constitutive arsenite oxidase expression detected in arsenic-hypertolerant Pseudomonas xanthomarina S11[J]. Res Microbiol, 2015, 166(3):205-214.
doi: 10.1016/j.resmic.2015.02.010 URL |
| [17] |
He M, Li X, Guo L, et al. Characterization and genomic analysis of chromate resistant and reducing Bacillus cereus strain SJ1[J]. BMC Microbiol, 2010, 10:221.
doi: 10.1186/1471-2180-10-221 URL |
| [18] | 车媛媛. 嗜铁钩端螺旋菌抗砷基因簇的生物信息学分析及改造[D]. 济南:山东大学, 2010. |
| Che YY. Bioinformatics analysis and mutation of Leptospirillum ferriphilum arsenic resistance system(ARS)cluster[D]. Jinan:Shandong University, 2010. | |
| [19] |
Chivers PT, Sauer RT. Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites[J]. J Biol Chem, 2000, 275(26):19735-19741.
pmid: 10787413 |
| [20] |
Xie P, Hao XL, Herzberg M, et al. Genomic analyses of metal resistance genes in three plant growth promoting bacteria of legume plants in Northwest mine tailings, China[J]. J Environ Sci, 2015, 27:179-187.
doi: 10.1016/j.jes.2014.07.017 URL |
| [21] |
Nies DH. Heavy metal-resistant bacteria as extremophiles:molecular physiology and biotechnological use of Ralstonia sp. CH34[J]. Extremophiles, 2000, 4(2):77-82.
pmid: 10805561 |
| [22] |
Liu P, Chen X, Huang Q, et al. The role of CzcRS two-component systems in the heavy metal resistance of Pseudomonas putida X4[J]. Int J Mol Sci, 2015, 16(8):17005-17017.
doi: 10.3390/ijms160817005 URL |
| [23] |
Serrato-Gamiño N, Salgado-Lora MG, Chávez-Moctezuma MP, et al. Analysis of the ars gene cluster from highly arsenic-resistant Burkholderia xenovorans LB400[J]. World J Microbiol Biotechnol, 2018, 34(10):1-10.
doi: 10.1007/s11274-017-2385-4 URL |
| [24] |
Cai L, Rensing C, Li XY, et al. Novel gene clusters involved in arsenite oxidation and resistance in two arsenite oxidizers:Achromobacter sp. SY8 and Pseudomonas sp. TS44[J]. Appl Microbiol Biotechnol, 2009, 83(4):715-725.
doi: 10.1007/s00253-009-1929-4 URL |
| [1] | 余洋, 刘天海, 刘理旭, 唐杰, 彭卫红, 陈阳, 谭昊. 羊肚菌菌种生产车间气溶胶微生物群落研究[J]. 生物技术通报, 2023, 39(5): 267-275. |
| [2] | 李会杰, 董莲华, 陈桂芳, 刘思渊, 杨佳怡, 杨靖亚. 食品中椰毒假单胞菌微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2023, 39(1): 127-136. |
| [3] | 贺丽娜, 冯源, 石慧敏, 叶建仁. 具有杀线活性马尾松内生细菌的筛选与鉴定[J]. 生物技术通报, 2022, 38(8): 159-166. |
| [4] | 史亚楠, 王德培, 王一川, 周昊, 薛鲜丽. 敲除msn2对米曲霉生长和发酵产曲酸的影响[J]. 生物技术通报, 2022, 38(8): 188-197. |
| [5] | 付雅丽, 彭万里, 林双君, 邓子新, 梁如冰. 香茅醇假单胞菌SJTE-3的短链脱氢酶SDR-X1的克隆及酶性质测定[J]. 生物技术通报, 2022, 38(3): 121-129. |
| [6] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [7] | 燕炯, 冯晨毅, 高学坤, 许祥, 杨佳敏, 陈朝阳. 基于CRISPR/Cas9技术构建Plin1基因敲除小鼠模型及表型分析[J]. 生物技术通报, 2022, 38(3): 173-180. |
| [8] | 胡珊, 梁卫驱, 黄皓, 徐匆, 罗华建, 胡楚维, 黄晓彦, 陈仕丽. 中药渣堆肥中解磷细菌的筛选、鉴定及其拮抗作用[J]. 生物技术通报, 2022, 38(3): 92-102. |
| [9] | 钟菁, 孙玲玲, 张姝, 蒙园, 支怡飞, 涂黎晴, 徐天鹏, 濮黎萍, 陆阳清. 应用CRISPR/Cas9技术敲除Mda5基因对新城疫及传染性法氏囊病毒复制的影响[J]. 生物技术通报, 2022, 38(11): 90-96. |
| [10] | 赵洋, 孙慧明, 林浩澎, 罗娉婷, 朱雅婷, 陈琼华, 舒琥. 一株安全高效的好氧反硝化菌Pseudomonas stutzeri DZ11的生物安全性及脱氮性能研究[J]. 生物技术通报, 2022, 38(10): 226-234. |
| [11] | 王海杰, 王成稷, 郭洋, 王云, 陈艳娟, 梁敏, 王珏, 龚慧, 沈如凌. 基于CRSIPR/Cas9技术构建凝血因子8基因敲除小鼠模型及表型验证[J]. 生物技术通报, 2022, 38(10): 273-280. |
| [12] | 王睿, 韩烈保. CRISPR/Cas9介导的二穗短柄草bdfls2敲除突变体的获得[J]. 生物技术通报, 2022, 38(1): 70-76. |
| [13] | 洪军, 卫夏怡, 吉冰洁, 叶延欣, 程天赐. 铜绿假单胞菌对鲎素耐药前后的差异表达基因及SNP变化研究[J]. 生物技术通报, 2021, 37(9): 191-202. |
| [14] | 蔡国磊, 陆小凯, 娄水珠, 杨海英, 杜刚. 芽孢杆菌LM基于全基因组的分类鉴定及抑菌原理的研究[J]. 生物技术通报, 2021, 37(8): 176-185. |
| [15] | 蒋成辉, 曾巧英, 王萌, 潘阳阳, 刘旭明, 尚天甜. CRISPR/Cas9构建srtA基因敲除的金黄色葡萄球菌[J]. 生物技术通报, 2020, 36(9): 253-265. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
