








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 256-263.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0550
张雨函1( ), 范熠1, 李婷婷1, 庞爽1, 刘为1, 白可喻2, 张西美1(
), 范熠1, 李婷婷1, 庞爽1, 刘为1, 白可喻2, 张西美1( )
)
收稿日期:2021-04-25
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:张雨函,女,硕士研究生,研究方向:农业水土环境;E-mail: 基金资助:
ZHANG Yu-han1( ), FAN Yi1, LI Ting-ting1, PANG Shuang1, LIU Wei1, BAI Ke-yu2, ZHANG Xi-mei1(
), FAN Yi1, LI Ting-ting1, PANG Shuang1, LIU Wei1, BAI Ke-yu2, ZHANG Xi-mei1( )
)
Received:2021-04-25
Published:2022-03-26
Online:2022-04-06
摘要:
获得高质量的微生物基因组DNA是进行复杂微生物群落宏基因组学研究的基础和难点。植物叶表是一个微生物多样性丰富的复杂生态系统,这些微生物群落可以调节叶片功能性状,影响植物的适应性。深入了解叶表微生物群落的基本结构和功能原理,有助于在促进植物生长和植物保护方面发挥重要的应用价值。由于叶表严苛的环境,导致富集叶表微生物难度较大,严重限制了高质量叶表微生物基因组DNA的提取。基于现有DNA提取方法,加入表面活性剂Silwet L-77进行前处理,同时循环利用洗脱液,加强叶表微生物的富集,以提高叶表微生物的获取量。结合商业试剂盒方法进行提取得到高纯度、高浓度的基因组DNA。经过质量控制和建库测序验证,DNA质量达到宏基组建库的要求。通过此方法可以提高叶表微生物分离和收集效率的方法,提高叶表微生物DNA提取成功率,为应用高通量测序技术研究叶表微生物组成和其他植物分子生物学研究提供参考。
张雨函, 范熠, 李婷婷, 庞爽, 刘为, 白可喻, 张西美. 基于宏基因组测序的植物叶表微生物富集及DNA提取方法[J]. 生物技术通报, 2022, 38(3): 256-263.
ZHANG Yu-han, FAN Yi, LI Ting-ting, PANG Shuang, LIU Wei, BAI Ke-yu, ZHANG Xi-mei. Microbial Enrichment on Leaf Surface and DNA Extraction Method Based on the Metagenomics Sequencing[J]. Biotechnology Bulletin, 2022, 38(3): 256-263.
| 样品名称 Sample name | 无菌水Sterile water | PBS+Silwet L-77 | ||||||
|---|---|---|---|---|---|---|---|---|
| DNA浓度DNA concentration/(ng·µL-1) | A260/A280 | 质量 Mass/µg | DNA浓度DNA concentration/(ng·µL-1) | A260/A280 | 质量 Mass/ µg | |||
| LC | 14.43±2.79 | 1.86 | 2.94 | 40.08±10.29 | 1.82 | 19.30 | ||
| PD | 13.61±7.52 | 1.91 | 1.44 | 74.25±5.77 | 1.96 | 19.85 | ||
| AF | 11.51±2.33 | 1.84 | 1.15 | 49.55±0.21 | 1.93 | 7.44 | ||
| IL | 19.51±8.13 | 1.89 | 1.95 | 64.42±13.03 | 1.93 | 34.97 | ||
| TL | 18.75±3.61 | 1.99 | 1.78 | 35.45±7.42 | 1.81 | 22.28 | ||
| AR | 18.14±3.87 | 1.97 | 1.81 | 44.93±31.67 | 1.96 | 30.31 | ||
| All | 15.99±4.95 | 1.93 | 1.63 | 51.44±18.10 | 1.86 | 22.36 | ||
表1 无菌水和PBS缓冲液+Silwet L-77表面活性剂对叶表微生物基因组DNA的提取效果
Table 1 Extraction effect of sterile water and PBS buffer solution +Silwet L-77 surfactant on microbial genomic DNA from leaf surface
| 样品名称 Sample name | 无菌水Sterile water | PBS+Silwet L-77 | ||||||
|---|---|---|---|---|---|---|---|---|
| DNA浓度DNA concentration/(ng·µL-1) | A260/A280 | 质量 Mass/µg | DNA浓度DNA concentration/(ng·µL-1) | A260/A280 | 质量 Mass/ µg | |||
| LC | 14.43±2.79 | 1.86 | 2.94 | 40.08±10.29 | 1.82 | 19.30 | ||
| PD | 13.61±7.52 | 1.91 | 1.44 | 74.25±5.77 | 1.96 | 19.85 | ||
| AF | 11.51±2.33 | 1.84 | 1.15 | 49.55±0.21 | 1.93 | 7.44 | ||
| IL | 19.51±8.13 | 1.89 | 1.95 | 64.42±13.03 | 1.93 | 34.97 | ||
| TL | 18.75±3.61 | 1.99 | 1.78 | 35.45±7.42 | 1.81 | 22.28 | ||
| AR | 18.14±3.87 | 1.97 | 1.81 | 44.93±31.67 | 1.96 | 30.31 | ||
| All | 15.99±4.95 | 1.93 | 1.63 | 51.44±18.10 | 1.86 | 22.36 | ||
| Df | F value | Pr(>F) | |
|---|---|---|---|
| 富集方法Enrichment method | 1 | 59.11 | P<0.05 |
| 植物种类Plant species | 5 | 1.71 | 0.21 |
| 富集方法:植物种类 Enrichment method:Plant species | 5 | 1.96 | 0.16 |
| 残差Residual | 12 | NA | NA |
表2 植物种类与不同富集方法对DNA浓度的影响
Table 2 Effects of plant species and different enrichment methods on DNA concentration
| Df | F value | Pr(>F) | |
|---|---|---|---|
| 富集方法Enrichment method | 1 | 59.11 | P<0.05 |
| 植物种类Plant species | 5 | 1.71 | 0.21 |
| 富集方法:植物种类 Enrichment method:Plant species | 5 | 1.96 | 0.16 |
| 残差Residual | 12 | NA | NA |
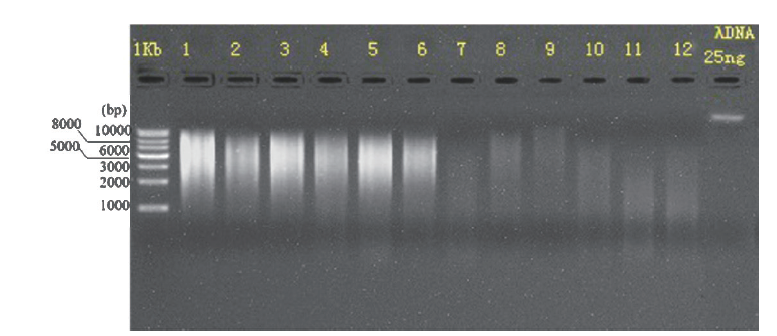
图2 六种植物叶表微生物DNA电泳图 1-6和7-12分别是羊草、叉分蓼、冷蒿、马蔺、披针叶黄华和野韭,1-6以PBS+ Silwet L-77为洗脱液,7-12以无菌水为洗脱液
Fig. 2 Microbial DNA electrophoresis patterns on the leaf surfaces of 6 plants 1-6 and 7-12 were L. chinensis(Trin.)Tzvel.,P. divaricatum L.,A. frigida Willd.,I. lactea Pall. var. chinensis(Fisch.)Koidz.,T. lanceolata R. Br. and A.ramosum L.. PBS+ Silwet L-77 as eluent for 1-6,and sterile water as eluent for 7-12,respectively
| 样品名称Sample name | Clean reads | Clean base/bp | Percent in raw reads/% | Percent in raw bases/% |
|---|---|---|---|---|
| LC | 84554592 | 12743415315 | 99.08±0.12 | 98.89±0.26 |
| PD | 76636682 | 11558890595 | 98.97±0.04 | 98.86±0.09 |
| AF | 70985020 | 10682348648 | 98.70±0.10 | 98.36±0.14 |
| IL | 77764022 | 11727532109 | 99.11±0.08 | 98.97±0.11 |
| TL | 87048063 | 13081919500 | 98.93±0.08 | 98.47±0.24 |
| AR | 80930005 | 12202864242 | 98.82±0.69 | 98.67±0.74 |
表3 宏基因组测序统计
Table 3 Metagenomics sequencing statistics
| 样品名称Sample name | Clean reads | Clean base/bp | Percent in raw reads/% | Percent in raw bases/% |
|---|---|---|---|---|
| LC | 84554592 | 12743415315 | 99.08±0.12 | 98.89±0.26 |
| PD | 76636682 | 11558890595 | 98.97±0.04 | 98.86±0.09 |
| AF | 70985020 | 10682348648 | 98.70±0.10 | 98.36±0.14 |
| IL | 77764022 | 11727532109 | 99.11±0.08 | 98.97±0.11 |
| TL | 87048063 | 13081919500 | 98.93±0.08 | 98.47±0.24 |
| AR | 80930005 | 12202864242 | 98.82±0.69 | 98.67±0.74 |
| [1] |
Zimmerman NB, Vitousek PM. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape[J]. PNAS, 2012, 109(32):13022-13027.
doi: 10.1073/pnas.1209872109 pmid: 22837398 |
| [2] |
Vorholt JA. Microbial life in the phyllosphere[J]. Nat Rev Microbiol, 2012, 10(12):828-840.
doi: 10.1038/nrmicro2910 pmid: 23154261 |
| [3] | 潘建刚, 呼庆, 齐鸿雁, 等. 叶际微生物研究进展[J]. 生态学报, 2011, 31(2):583-592. |
| Pan JG, Hu Q, Qi HY, et al. Advance in the research of phyllospheric microorganism[J]. Acta Ecol Sin, 2011, 31(2):583-592. | |
| [4] |
Lindow SE, Brandl MT. Microbiology of the phyllosphere[J]. Appl Environ Microbiol, 2003, 69(4):1875-1883.
doi: 10.1128/AEM.69.4.1875-1883.2003 URL |
| [5] |
Yang CH, Crowley DE, Borneman J, et al. Microbial phyllosphere populations are more complex than previously realized[J]. PNAS, 2001, 98(7):3889-3894.
pmid: 11274410 |
| [6] |
Chaudhry V, Runge P, Sengupta P, et al. Shaping the leaf microbiota:plant-microbe-microbe interactions[J]. J Exp Bot, 2021, 72(1):36-56.
doi: 10.1093/jxb/eraa417 pmid: 32910810 |
| [7] | Soni R, Kumar V, Suyal DC, et al. Metagenomics of plant rhizosphere microbiome[M]// Singh RP, Kothari R, Koringa PG, et al. Understanding host-microbiome interactions-an omics approach. Springer Singapore:Singapore, 2017:193-205. |
| [8] | 张泽生, 王春龙, 刘清岱, 等. 扩增子测序技术分析红茶菌中优势微生物的研究[J]. 食品研究与开发, 2016, 37(16):185-188. |
| Zhang ZS, Wang CL, Liu QD, et al. Study on analysis of the dominant microorganisms in kombucha by the technology of amplicon sequencing[J]. Food Res Dev, 2016, 37(16):185-188. | |
| [9] |
田李, 张颖, 赵云峰. 新一代测序技术的发展和应用[J]. 生物技术通报, 2015, 31(11):1-8.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.11.003 |
|
Tian L, Zhang Y, Zhao YF. The next generation sequencing technology and its applications[J]. Biotechnol Bull, 2015, 31(11):1-8.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.11.003 |
|
| [10] | 魏子艳, 金德才, 邓晔. 环境微生物宏基因组学研究中的生物信息学方法[J]. 微生物学通报, 2015, 42(5):890-901. |
| Wei ZY, Jin DC, Deng Y. Bioinformatics tools and applications in the study of environmental microbial metagenomics[J]. Microbiol China, 2015, 42(5):890-901. | |
| [11] |
Zhang XM, Johnston ER, Barberán A, et al. Decreased plant productivity resulting from plant group removal experiment constrains soil microbial functional diversity[J]. Glob Change Biol, 2017, 23(10):4318-4332.
doi: 10.1111/gcb.2017.23.issue-10 URL |
| [12] |
Laforest-Lapointe I, Whitaker BK. Decrypting the phyllosphere microbiota:progress and challenges[J]. Am J Bot, 2019, 106(2):171-173.
doi: 10.1002/ajb2.1229 pmid: 30726571 |
| [13] |
Handelsman J. Metagenomics:application of genomics to uncultured microorganisms[J]. Microbiol Mol Biol Rev, 2004, 68(4):669-685.
doi: 10.1128/MMBR.68.4.669-685.2004 URL |
| [14] |
Wang HX, Geng ZL, Zeng Y, et al. Enriching plant microbiota for a metagenomic library construction[J]. Environ Microbiol, 2008, 10(10):2684-2691.
doi: 10.1111/j.1462-2920.2008.01689.x URL |
| [15] | National Research Council(US)Committee on Metagenomics Challenges. The new science of metagenomics[M]. Washington DC:National Academies Press, 2007. |
| [16] |
Bag S, Saha B, Mehta O, et al. An improved method for high quality metagenomics DNA extraction from human and environmental samples[J]. Sci Rep, 2016, 6:26775.
doi: 10.1038/srep26775 URL |
| [17] |
Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms:cutting the Gordian knot[J]. Genome Biol, 2005, 6(8):229.
doi: 10.1186/gb-2005-6-8-229 URL |
| [18] |
赵玲云, 范东颖, 李燕芳, 等. 枝干树皮宏基因组DNA的提取[J]. 生物技术通报, 2016, 32(1):74-79.
doi: 10.13560/j.cnki.biotech.bull.1985.2016.01.018 |
| Zhao LY, Fan DY, Li YF, et al. The extraction of metagenom DNA in branch bark[J]. Biotechnol Bull, 2016, 32(1):74-79. | |
| [19] | 周育, 乔雄梧, 王静, 等. 植物叶际微生物提取方法研究[J]. 植物研究, 2006, 26(2):2233-2237. |
| Zhou Y, Qiao XW, Wang J, et al. Extraction methods of microorganisms from phyllosphere[J]. Bull Bot Res, 2006, 26(2):2233-2237. | |
| [20] |
Schreiber L, Krimm U, Knoll D, et al. Plant-microbe interactions:identification of epiphytic bacteria and their ability to alter leaf surface permeability[J]. New Phytol, 2005, 166(2):589-594.
pmid: 15819920 |
| [21] | 赵裕栋, 周俊, 何璟. 土壤微生物总DNA提取方法的优化[J]. 微生物学报, 2012, 52(9):1143-1150. |
| Zhao YD, Zhou J, He J. Optimization of soil microbial DNA isolation[J]. Acta Microbiol Sin, 2012, 52(9):1143-1150. | |
| [22] | 阳静, 张静, 邹伟, 等. 环境微生物DNA提取方法研究进展[J]. 食品与机械, 2017, 33(3):207-210, 215. |
| Yang J, Zhang J, Zou W, et al. Progress of study on extraction methods of environmental microbial DNA[J]. Food Mach, 2017, 33(3):207-210, 215. | |
| [23] |
Kembel SW, O’Connor TK, Arnold HK, et al. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest[J]. PNAS, 2014, 111(38):13715-13720.
doi: 10.1073/pnas.1216057111 URL |
| [24] | 逄森, 袁会珠, 李永平, 等. 表面活性剂silwet408提高药液在蔬菜叶片上润湿性能的研究[J]. 农药科学与管理, 2005, 26(7):22-25, 19. |
| Pang S, Yuan HZ, Li YP, et al. Study on the wetting property of surfactant Silwet408 on vegetable leaf surface[J]. Pestic Sci Adm, 2005, 26(7):22-25, 19. | |
| [25] |
Luo L, Zhang Z, Wang P, et al. Variations in phyllosphere microbial community along with the development of angular leaf-spot of cucumber[J]. AMB Express, 2019, 9(1):76.
doi: 10.1186/s13568-019-0800-y URL |
| [26] | 张丽, 郭佩瑶, 张春华, 等. 农杆菌介导的eGFP基因转化瓜叶菊技术体系的建立[J]. 分子植物育种, 2020, 18(19):6350-6358. |
| Zhang L, Guo PY, Zhang CH, et al. Establishment of Agrobacterium tumefaciens-mediated genetic transformation system of eGFP gene in Pericallis hybrida[J]. Mol Plant Breed, 2020, 18(19):6350-6358. | |
| [27] | 罗世地, 莫春生, 夏娇, 等. Tween 80分子在气-液界面上的吸附动力学[J]. 湛江海洋大学学报, 2005, 25(4):49-53. |
| Luo SD, Mo CS, Xia J, et al. Adsorptive kinetics of aqueous tween 80 solutionsat the air-water interface[J]. J Zhanjiang Ocean Univ, 2005, 25(4):49-53. | |
| [28] |
Delmotte N, Knief C, Chaffron S, et al. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria[J]. PNAS, 2009, 106(38):16428-16433.
doi: 10.1073/pnas.0905240106 URL |
| [29] |
Bulgarelli D, Rott M, Schlaeppi K, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota[J]. Nature, 2012, 488(7409):91-95.
doi: 10.1038/nature11336 URL |
| [30] |
Knief C, Delmotte N, Chaffron S, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice[J]. Isme J, 2012, 6(7):1378-1390.
doi: 10.1038/ismej.2011.192 URL |
| [31] |
Fadiji AE, Babalola OO. Metagenomics methods for the study of plant-associated microbial communities:a review[J]. J Microbiol Methods, 2020, 170:105860.
doi: 10.1016/j.mimet.2020.105860 URL |
| [32] | Knief C. Analysis of plant microbe interactions in the era of next generation sequencing technologies[J]. Front Plant Sci, 2014, 5:216. |
| [33] | 高爽, 刘笑尘, 董铮, 等. 叶际微生物及其与外界互作的研究进展[J]. 植物科学学报, 2016, 34(4):654-661. |
| Gao S, Liu XC, Dong Z, et al. Advance of phyllosphere microorganisms and their interaction with the outside environment[J]. Plant Sci J, 2016, 34(4):654-661. |
| [1] | 申恒, 刘思慧, 李跃, 李敬涛, 梁文星. 一种用于PCR的番茄DNA快速粗提方法[J]. 生物技术通报, 2022, 38(6): 74-80. |
| [2] | 张淼, 孙祥瑞, 徐春明. 单细胞RNA测序数据分析方法研究进展[J]. 生物技术通报, 2021, 37(1): 52-59. |
| [3] | 杨秀清, 陈彦梅, 吴瑞薇, 王保玉, 韩作颖. 煤地质环境微生物总基因组DNA提取方法的优化[J]. 生物技术通报, 2018, 34(9): 177-183. |
| [4] | 李海洋, 王飞, 雷红涛, 张璇, 陈珂. 硅羟基磁珠的制备及全基因组DNA提取优化[J]. 生物技术通报, 2017, 33(6): 223-229. |
| [5] | 徐宗昌, 王萌, 孔英珍. 烟草碱法提取基因组DNA的改进[J]. 生物技术通报, 2017, 33(2): 59-65. |
| [6] | 张红利,贾鑫磊,刘建霞,张永芳. 保存方式和回软温度对蜜蜂标本DNA提取的影响[J]. 生物技术通报, 2016, 32(7): 250-256. |
| [7] | 周海兰,李绍鹏,李卫亮,贺军虎,包冬红,李茂富. 油梨基因组DNA提取、SSR-PCR反应体系优化及引物筛选[J]. 生物技术通报, 2016, 32(4): 143-150. |
| [8] | 赵玲云, 范东颖, 李燕芳, 颜霞, 黄丽丽. 枝干树皮宏基因组DNA的提取[J]. 生物技术通报, 2016, 32(1): 74-79. |
| [9] | 张振华,鲍王波,余冉,王长永,刘燕. 鹌鹑肠道微生物PCR-DGGE方法的优化[J]. 生物技术通报, 2015, 31(1): 73-78. |
| [10] | 戴恬美, 吕志创, 万方浩. 烟粉虱全基因组DNA四种提取方法的比较[J]. 生物技术通报, 2014, 0(8): 70-75. |
| [11] | 李进波, 盛婧, 李想, 潘良文, 吕蓉, 杨捷琳. 五种DNA提取方法对鱼加工制品DNA提取效果的比较[J]. 生物技术通报, 2014, 0(4): 43-49. |
| [12] | 权俊萍;贾晓鹰;戴丽娜;肖俊辉;田琴;牛攀新;吕国华;. 薰衣草高质量DNA提取及ISSR-PCR多重化体系的建立[J]. , 2012, 0(05): 151-157. |
| [13] | 武强;吕志创;万方浩;李照会;. 三种提取苹果绵蚜基因组DNA方法的比较[J]. , 2012, 0(01): 70-73. |
| [14] | 蔡翠霞;肖维威;康文杰;周琳华;吴永彬;郑远金;马文丽;. 改良Chelex-100法快速提取转基因农产品DNA[J]. , 2011, 0(10): 210-214. |
| [15] | 代文娟;唐文秀;邓涛;丁莉;骆文华;黄仕训;. 狭叶坡垒基因组总DNA的提取和纯化方法研究[J]. , 2011, 0(07): 101-105. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||