








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 115-127.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0270
收稿日期:2023-03-24
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
支添添,女,博士,讲师,研究方向:甘蓝型油菜和拟南芥酪氨酸缺陷突变体sscd1抗病机制;E-mail: 190055@jxycu.edu.cn; 基金资助:
ZHI Tian-tian1( ), ZHOU Zhou1, CHEN Ji-peng1, HAN Cheng-yun2
), ZHOU Zhou1, CHEN Ji-peng1, HAN Cheng-yun2
Received:2023-03-24
Published:2023-10-26
Online:2023-11-28
摘要:
克隆甘蓝型油菜(Brassica napus L.)酪氨酸代谢关键基因FAH,对其进行功能验证和表达分析,为进一步解析FAH在甘蓝型油菜中的作用和功能提供理论依据。以甘蓝型油菜‘westar’为试材,克隆与拟南芥AtFAH同源性最高的甘蓝型油菜FAH基因BnaA06g38260D(BnaA06FAH)和BnaC05g49430D(BnaC05FAH),通过生物信息学分析其亲缘关系,构建过表达载体转化拟南芥突变体sscd1进行功能验证。克隆BnaA06FAH和BnaC05FAH启动子序列,利用PlantCare在线数据库分析启动子调控元件,构建启动子和GUS的融合载体,通过GUS组织化学染色分析其表达模式。结果显示,BnaA06FAH和BnaC05FAH与AtFAH的氨基酸序列相似性分别为93.11%和92.40%,2个基因过表达都可以完全抑制拟南芥突变体sscd1在短日照下模拟病斑的形成,暗示BnaA06FAH和BnaC05FAH都与AtFAH功能相似。BnaA06FAH和BnaC05FAH启动子除具有所必需的TATA-box和CAAT-box等基本顺式作用元件外,都含有多个与光诱导、激素响应和逆境胁迫响应元件以及多种与抗病相关的顺式作用元件,但与BnaA06FAH相比,BnaC05FAH与拟南芥AtFAH相同的顺式作用元件更多;GUS活性检测表明,BnaC05FAH启动子驱动的GUS基因的表达比BnaA06FAH强,并且两者驱动表达的组织部位不完全相同。因此,BnaA06FAH和BnaC05FAH启动子的作用部位和作用强度都存在明显差异。BnaA06FAH和BnaC05FAH都能够调控拟南芥sscd1模拟病斑的形成,但两者启动子驱动下游基因的强度和部位不同。
支添添, 周舟, 陈纪鹏, 韩成云. 甘蓝型油菜酪氨酸代谢关键基因FAH的克隆、功能鉴定和表达分析[J]. 生物技术通报, 2023, 39(10): 115-127.
ZHI Tian-tian, ZHOU Zhou, CHEN Ji-peng, HAN Cheng-yun. Cloning, Functional Identification and Expression Analysis of FAH, a Key Gene for Tyrosine Metabolism in Brassica napus L.[J]. Biotechnology Bulletin, 2023, 39(10): 115-127.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| BnaC05FAH-ty-F | AACCTACCACTGCCCTTT |
| BnaC05FAH-ty-R | TTTCCACCAACTTCCTGC |
| BnaA06FAH-ty-F | ACCATACATCATCCGTTT |
| BnaA06FAH-ty-R | TCAAGGCAGTGAAGGTAAAA |
| BnaC05FAH-Xba I -F | GCTCTAGACCATGGCGTTGCTCAAGTCTTT |
| BnaC05FAH-Sac I -R | CGAGCTCCATCAAGGCAGTGAAGGTAAAA |
| BnaA06FAH-Xba I -F | GCTCTAGAATGGCGTTGCTCAAGTCTTTCG |
| BnaA06FAH-Sac I -R | CGAGCTCTCAAGGCAGTGAAGGTAAAAT |
| BnaC05FAHpro-Xba I -F | GCTCTAGATTAAGGTAATGCTTTGATTCG |
| BnaC05FAHpro-Bgl II -R | GGAAGATCTGGACGATAAACAAACAGAT |
| BnaA06FAHpro-Xba I -F | GCTCTAGAAGTTCCCTAACGTTGTCCTCACTTT |
| BnaA06FAHpro-Bgl II -R | GGAAGATCTTGTATGACGAAGTTTCCAAAGC |
| qBnaC05FAH-F | AACCTACCACTGCCCTTT |
| qBnaC05FAH-R | CGTATGACGATGTTTCCG |
| qBnaA06FAH-F | TTTGGAAACTTCACCGAG |
| qBnaA06FAH-R | GAGTGTGGAGCAACATCG |
| ACTIN2-F | AGCACTTGCACCAAGCAGCATG |
| ACTIN2-R | ACGATTCCTGGACCTGCCTCATC |
表1 试验所用引物
Table 1 Primers used in the experiment
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| BnaC05FAH-ty-F | AACCTACCACTGCCCTTT |
| BnaC05FAH-ty-R | TTTCCACCAACTTCCTGC |
| BnaA06FAH-ty-F | ACCATACATCATCCGTTT |
| BnaA06FAH-ty-R | TCAAGGCAGTGAAGGTAAAA |
| BnaC05FAH-Xba I -F | GCTCTAGACCATGGCGTTGCTCAAGTCTTT |
| BnaC05FAH-Sac I -R | CGAGCTCCATCAAGGCAGTGAAGGTAAAA |
| BnaA06FAH-Xba I -F | GCTCTAGAATGGCGTTGCTCAAGTCTTTCG |
| BnaA06FAH-Sac I -R | CGAGCTCTCAAGGCAGTGAAGGTAAAAT |
| BnaC05FAHpro-Xba I -F | GCTCTAGATTAAGGTAATGCTTTGATTCG |
| BnaC05FAHpro-Bgl II -R | GGAAGATCTGGACGATAAACAAACAGAT |
| BnaA06FAHpro-Xba I -F | GCTCTAGAAGTTCCCTAACGTTGTCCTCACTTT |
| BnaA06FAHpro-Bgl II -R | GGAAGATCTTGTATGACGAAGTTTCCAAAGC |
| qBnaC05FAH-F | AACCTACCACTGCCCTTT |
| qBnaC05FAH-R | CGTATGACGATGTTTCCG |
| qBnaA06FAH-F | TTTGGAAACTTCACCGAG |
| qBnaA06FAH-R | GAGTGTGGAGCAACATCG |
| ACTIN2-F | AGCACTTGCACCAAGCAGCATG |
| ACTIN2-R | ACGATTCCTGGACCTGCCTCATC |
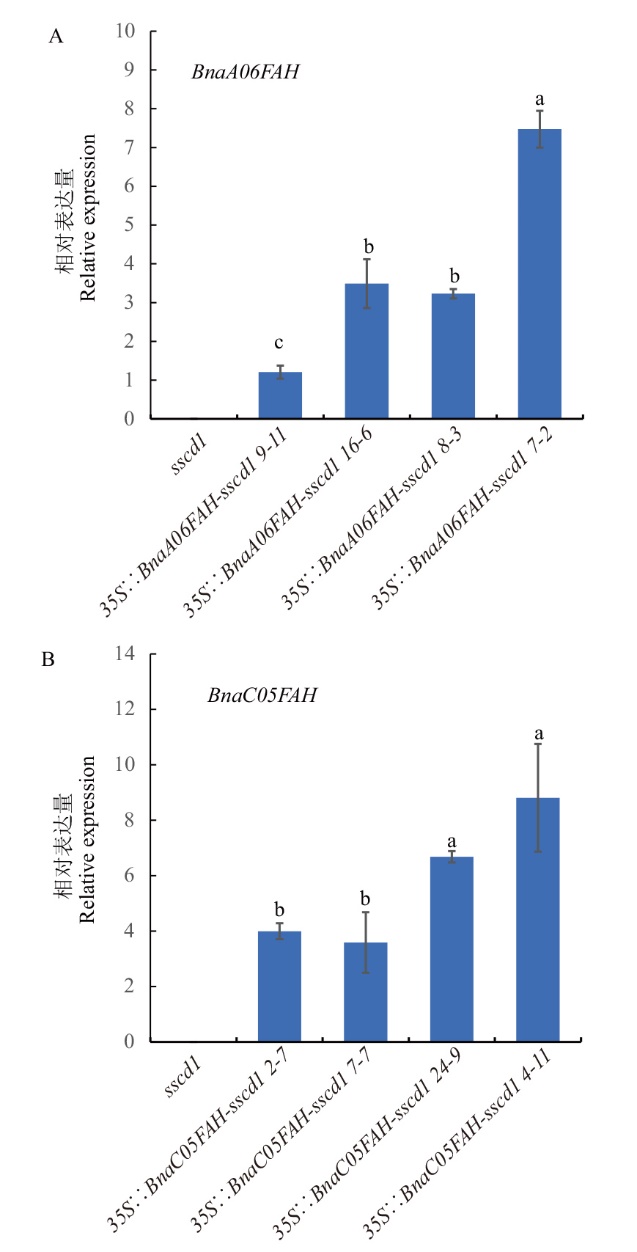
图3 过表达拟南芥株系中BnaA06FAH和BnaC05FAH的相对表达量 不同小写字母表示差异显著(P<0.05)
Fig 3 Relative expressions of BnaA06FAH and BnaC05-FAH in overexpressed A. thaliana line Different lowercases indicate significant difference(P<0.05)

图4 短日照下过表达BnaA06FAH和BnaC05FAH抑制拟南芥突变体sscd1的模拟病斑 A:短日照下生长10 d的幼苗;B:长日照下生长20 d转入短日照3 d的莲座叶
Fig. 4 Overexpressions of BnaA06FAH and BnaC05FAH repress the simulated disease spot in Arabidopsis mutant sscd1 under short-day condition A: Seedlings grown for 10 d under short day condition; B: rosettes grow under long day condition for 20 d then transferred to short day for 3 d
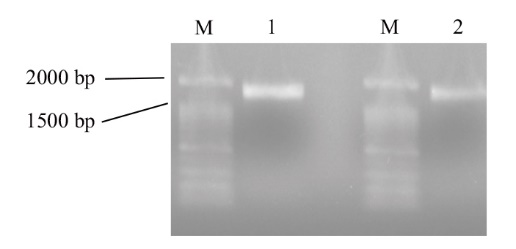
图5 BnaA06FAH和BnaC05FAH启动子的PCR扩增产物
Fig. 5 PCR products of BnaA06FAH and BnaC05FAH promotors M: DNA marker; 1: BnaA06FAH promotor; 2: BnaC05FAH promotor
| 基因 Gene | 顺式作用元件 Cis-acting element | 核心序列 Core sequence | 生物学功能 Biological function |
|---|---|---|---|
| BnaC05FAH BnaA06FAH AtFAH | CAAT-box | CCAAT/CAAT/CAAAT | 启动子和增强子区的一般顺式作用元件 Common cis-acting element in promoter and enhancer regions |
| TATA-box | ATTATA/TATACA/TATAAGAA | 转录起始-30核心启动子元件 Core promoter element around -30 of transcription start | |
| as-1 | TGACG | 胁迫响应元件Stress responsive element | |
| CGTCA-motif | CGTCA | 参与茉莉酸甲酯响应的顺式作用元件 Cis-acting regulatory element involved in the MeJA-responsiveness | |
| GT1-motif | GGTTAA | 光响应元件Light responsive element | |
| O2-site | GTTGACGTGA | 参与玉米醇溶蛋白代谢调控的顺式作用元件 Cis-acting regulatory element involved in zein metabolism regulation | |
| STRE | AGGGG | 胁迫响应元件Stress responsive element | |
| TCT-motif | TCTTAC | 部分光响应元件Part of a light responsive element | |
| TGACG-motif | TGACG | 参与茉莉酸甲酯响应的顺式作用元件 Cis-acting regulatory element involved in the MeJA-responsiveness | |
| BnaA06FAH AtFAH | ABRE | ACGTG/CACGTG | 脱落酸响应元件Abscisic acid responsive element |
| ABRE3a | TACGTG | 脱落酸响应元件Abscisic acid responsive element | |
| ABRE4 | CACGTA | 脱落酸响应元件Abscisic acid responsive element | |
| DRE core | GCCGAC | 脱水响应元件Dehydration responsive element | |
| G-box | TACGTG | 光响应元件Light responsive element | |
| Sp1 | GGGCGG | 光响应元件Light responsive element | |
| WRE3 | CCACCT | 创伤响应元件Wounding responsive element |
表2 BnaC05FAH、BnaA06FAH和AtFAH启动子区共同顺式作用元件
Table 2 Common cis-acting elements in BnaC05FAH, BnaA06FAH and AtFAH promoter regions
| 基因 Gene | 顺式作用元件 Cis-acting element | 核心序列 Core sequence | 生物学功能 Biological function |
|---|---|---|---|
| BnaC05FAH BnaA06FAH AtFAH | CAAT-box | CCAAT/CAAT/CAAAT | 启动子和增强子区的一般顺式作用元件 Common cis-acting element in promoter and enhancer regions |
| TATA-box | ATTATA/TATACA/TATAAGAA | 转录起始-30核心启动子元件 Core promoter element around -30 of transcription start | |
| as-1 | TGACG | 胁迫响应元件Stress responsive element | |
| CGTCA-motif | CGTCA | 参与茉莉酸甲酯响应的顺式作用元件 Cis-acting regulatory element involved in the MeJA-responsiveness | |
| GT1-motif | GGTTAA | 光响应元件Light responsive element | |
| O2-site | GTTGACGTGA | 参与玉米醇溶蛋白代谢调控的顺式作用元件 Cis-acting regulatory element involved in zein metabolism regulation | |
| STRE | AGGGG | 胁迫响应元件Stress responsive element | |
| TCT-motif | TCTTAC | 部分光响应元件Part of a light responsive element | |
| TGACG-motif | TGACG | 参与茉莉酸甲酯响应的顺式作用元件 Cis-acting regulatory element involved in the MeJA-responsiveness | |
| BnaA06FAH AtFAH | ABRE | ACGTG/CACGTG | 脱落酸响应元件Abscisic acid responsive element |
| ABRE3a | TACGTG | 脱落酸响应元件Abscisic acid responsive element | |
| ABRE4 | CACGTA | 脱落酸响应元件Abscisic acid responsive element | |
| DRE core | GCCGAC | 脱水响应元件Dehydration responsive element | |
| G-box | TACGTG | 光响应元件Light responsive element | |
| Sp1 | GGGCGG | 光响应元件Light responsive element | |
| WRE3 | CCACCT | 创伤响应元件Wounding responsive element |
| 基因 Gene | 顺式元件 Cis-acting element | 序列 Sequence(5'-3') | 生物学功能 Biological function |
|---|---|---|---|
| BnaC05FAH | AC-I | (T/C)C(T/C)(C/T)ACC(T/C)ACC | 木质部特异性表达顺式元件 Cis-acting regulatory element of xylem specific expression |
| AE-box | AGAAACAA | 光响应元件的一部分Part of a light responsive element | |
| AT1-motif | AATTATTTTTTATT | 光响应元件的一部分Part of a light responsive element | |
| Box 4 | ATTAAT | 参与光响应的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness | |
| ERE | ATTTTAAA | 乙烯响应元件Ethylene responsive element | |
| GA-motif | ATAGATAA | 光响应元件的一部分Part of a light responsive element | |
| WUN-motif | CAATTACAT | 创伤响应元件Wounding responsive element | |
| MRE | AACCTAA | 参与光响应的MYB结合位点 MYB binding site involved in light responsiveness | |
| BnaA06FAH | A-box | CCGTCC | 瞬时顺式调节元件Instantaneous cis-regulating element |
| AAGAA-motif | gGTAAAGAAA | 胚乳发育相关顺式作用元件 Cis-acting regulatory element involved in endosperm development | |
| GARE-motif | TCTGTTG | 赤霉素响应元件Gibberellin responsive element | |
| TGA-element | TGACGTAA | 生长素响应元件Auxin responsive element | |
| W-box | (T)TGAC(C/T) | 病原诱导相关基因的顺式作用元件 Cis-acting regulatory element of pathogen-induction-related genes | |
| AtFAH | ATCT-motif | AATCTAATCC | 参与光响应的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness |
| circadian | CAAAGATATC | 参与昼夜节律控制的顺式作用调节元件 Cis-acting regulatory element involved in circadian rhythm control | |
| G-Box | CACGTG | 参与光反应的顺式作用调节元件 Cis-acting regulatory element involved in light response |
表3 BnaC05FAH、BnaA06FAH和AtFAH启动子区包含的不同顺式作用元件
Table 3 Different cis-acting elements in BnaC05FAH, BnaA06FAH and AtFAH promoter regions
| 基因 Gene | 顺式元件 Cis-acting element | 序列 Sequence(5'-3') | 生物学功能 Biological function |
|---|---|---|---|
| BnaC05FAH | AC-I | (T/C)C(T/C)(C/T)ACC(T/C)ACC | 木质部特异性表达顺式元件 Cis-acting regulatory element of xylem specific expression |
| AE-box | AGAAACAA | 光响应元件的一部分Part of a light responsive element | |
| AT1-motif | AATTATTTTTTATT | 光响应元件的一部分Part of a light responsive element | |
| Box 4 | ATTAAT | 参与光响应的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness | |
| ERE | ATTTTAAA | 乙烯响应元件Ethylene responsive element | |
| GA-motif | ATAGATAA | 光响应元件的一部分Part of a light responsive element | |
| WUN-motif | CAATTACAT | 创伤响应元件Wounding responsive element | |
| MRE | AACCTAA | 参与光响应的MYB结合位点 MYB binding site involved in light responsiveness | |
| BnaA06FAH | A-box | CCGTCC | 瞬时顺式调节元件Instantaneous cis-regulating element |
| AAGAA-motif | gGTAAAGAAA | 胚乳发育相关顺式作用元件 Cis-acting regulatory element involved in endosperm development | |
| GARE-motif | TCTGTTG | 赤霉素响应元件Gibberellin responsive element | |
| TGA-element | TGACGTAA | 生长素响应元件Auxin responsive element | |
| W-box | (T)TGAC(C/T) | 病原诱导相关基因的顺式作用元件 Cis-acting regulatory element of pathogen-induction-related genes | |
| AtFAH | ATCT-motif | AATCTAATCC | 参与光响应的保守DNA模块的一部分 Part of a conserved DNA module involved in light responsiveness |
| circadian | CAAAGATATC | 参与昼夜节律控制的顺式作用调节元件 Cis-acting regulatory element involved in circadian rhythm control | |
| G-Box | CACGTG | 参与光反应的顺式作用调节元件 Cis-acting regulatory element involved in light response |
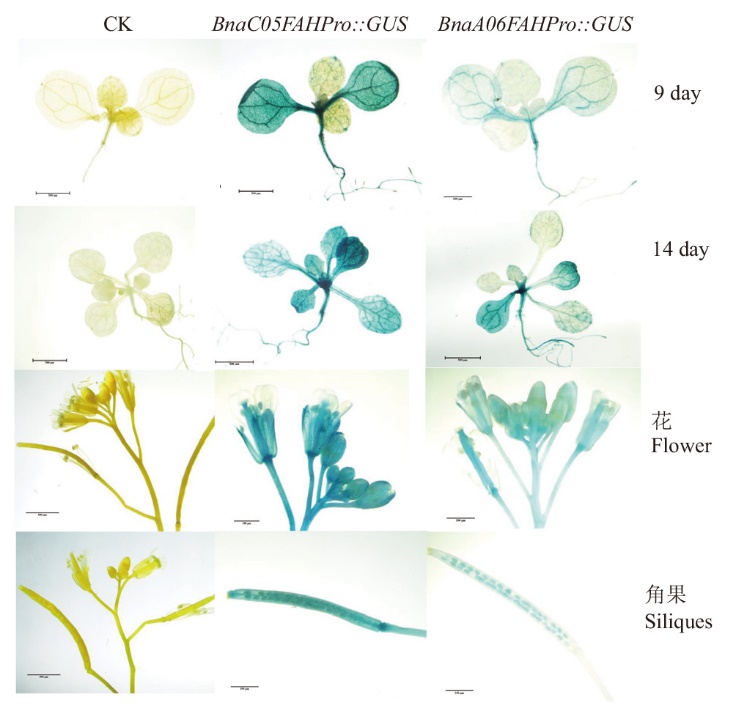
图7 甘蓝型油菜BnaC05FAH、BnaA06FAH启动子驱动GUS在拟南芥中的表达
Fig. 7 GUS expressions driven by BnaC05FAH and BnaA-06FAH promoters from B. napus in Arabidopsis seedlings
| [1] |
Lindblad B, Lindstedt S, Steen G. On the enzymic defects in hereditary tyrosinemia[J]. Proc Natl Acad Sci USA, 1977, 74(10): 4641-4645.
doi: 10.1073/pnas.74.10.4641 pmid: 270706 |
| [2] |
Ruppert S, Kelsey G, Schedl A, et al. Deficiency of an enzyme of tyrosine metabolism underlies altered gene expression in newborn liver of lethal albino mice[J]. Genes Dev, 1992, 6(8): 1430-1443.
doi: 10.1101/gad.6.8.1430 URL |
| [3] |
Grompe M al-Dhalimy M, Finegold M, et al. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice[J]. Genes Dev, 1993, 7(12a): 2298-2307.
doi: 10.1101/gad.7.12a.2298 URL |
| [4] |
Lock EA, Gaskin P, Ellis MK, et al. Tissue distribution of 2-(2-nitro-4-trifluoromethylbenzoyl)cyclohexane-1-3-dione(NTBC): effect on enzymes involved in tyrosine catabolism and relevance to ocular toxicity in the rat[J]. Toxicol Appl Pharmacol, 1996, 141(2): 439-447.
doi: 10.1006/taap.1996.0310 URL |
| [5] |
Sparnins VL, Chapman PJ. Catabolism of L-tyrosine by the homoprotocatechuate pathway in gram-positive bacteria[J]. J Bacteriol, 1976, 127(1): 362-266.
doi: 10.1128/jb.127.1.362-366.1976 pmid: 931949 |
| [6] |
Dixon DP, Edwards R. Enzymes of tyrosine catabolism in Arabidopsis thaliana[J]. Plant Sci, 2006, 171(3): 360-366.
doi: 10.1016/j.plantsci.2006.04.008 URL |
| [7] |
Hildebrandt T, Nunes Nesi A, Araújo W, et al. Amino acid catabolism in plants[J]. Mol Plant, 2015, 8(11): 1563-1579.
doi: 10.1016/j.molp.2015.09.005 pmid: 26384576 |
| [8] |
Xu JJ, Fang X, Li CY, et al. General and specialized tyrosine metabolism pathways in plants[J]. aBIOTECH, 2020, 1(2): 97-105.
doi: 10.1007/s42994-019-00006-w |
| [9] |
Schenck CA, Maeda HA. Tyrosine biosynthesis, metabolism, and catabolism in plants[J]. Phytochemistry, 2018, 149: 82-102.
doi: S0031-9422(18)30034-7 pmid: 29477627 |
| [10] |
Jin X, Chen X, et al. Imbalance of tyrosine by modulating TyrA arogenate dehydrogenases impacts growth and development of Arabidopsis thaliana[J]. Plant J, 2019, 97(5): 901-922.
doi: 10.1111/tpj.2019.97.issue-5 URL |
| [11] |
Han CY, Ren CM, Zhi TT, et al. Disruption of fumarylacetoacetate hydrolase causes spontaneous cell death under short-day conditions in Arabidopsis[J]. Plant Physiol, 2013, 162(4): 1956-1964.
doi: 10.1104/pp.113.216804 URL |
| [12] |
Zhou Z, Zhi T, Han C, et al. Cell death resulted from loss of fumarylacetoacetate hydrolase in Arabidopsis is related to phytohormone jasmonate but not salicylic acid[J]. Sci Rep, 2020, 10(1): 13714.
doi: 10.1038/s41598-020-70567-0 pmid: 32792583 |
| [13] |
Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants[J]. Annu Rev Plant Biol, 2012, 63: 73-105.
doi: 10.1146/annurev-arplant-042811-105439 pmid: 22554242 |
| [14] |
Singh S, Mishra VK, Kharwar RN, et al. Genetic characterization for lesion mimic and other traits in relation to spot blotch resistance in spring wheat[J]. PLoS One, 2020, 15(10): e0240029.
doi: 10.1371/journal.pone.0240029 URL |
| [15] |
Yu Y, Zhang Q, Sun S, et al. Upregulated expression of respiratory burst oxidase homolog d underlies lesion-mimic phenotype in dark-treated Arabidopsis pheide a oxygenase mutant leaves[J]. Planta, 2022, 255(6): 110.
doi: 10.1007/s00425-022-03895-2 |
| [16] |
Mu XH, Li JK, Dai ZZ, et al. Commonly and specifically activated defense responses in maize disease lesion mimic mutants revealed by integrated transcriptomics and metabolomics analysis[J]. Front Plant Sci, 2021, 12: 638792.
doi: 10.3389/fpls.2021.638792 URL |
| [17] | Zhao Y, Xu W, Wang L, et al. A maize necrotic leaf mutant caused by defect of coproporphyrinogen III oxidase in the porphyrin pathway[J]. Genes: Basel, 2022, 13(2): 272. |
| [18] |
Kang SG, Lee KE, Singh M, et al. Rice Lesion Mimic Mutants(LMM): The current understanding of genetic mutations in the failure of ROS scavenging during lesion formation[J]. Plants, 2021, 10(8): 1598.
doi: 10.3390/plants10081598 URL |
| [19] |
Sindhu A, Janick-Buckner D, Buckner B, et al. Propagation of cell death in dropdead1, a sorghum ortholog of the maize lls1 mutant[J]. PLoS One, 2018, 13(9): e0201359.
doi: 10.1371/journal.pone.0201359 URL |
| [20] |
Wang J, Ye BQ, Yin JJ, et al. Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice[J]. Plant Physiol Biochem, 2015, 97: 44-51.
doi: 10.1016/j.plaphy.2015.09.001 URL |
| [21] |
Yang S, Hua JA. haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis[J]. Plant Cell, 2004, 16: 1060-1071.
doi: 10.1105/tpc.020479 URL |
| [22] |
Gou M, Su N, Zheng J, et al. An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis[J]. Plant J, 2009, 60: 757-770.
doi: 10.1111/tpj.2009.60.issue-5 URL |
| [23] |
Liu Q, Ning Y, Zhang Y, et al. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice[J]. Plant Cell, 2017, 29: 345-359.
doi: 10.1105/tpc.16.00650 URL |
| [24] |
Wolter M, Hollricher K, Salamini F, et al. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype[J]. Mol Genet Genomics, 1993, 239: 122-128.
doi: 10.1007/BF00281610 |
| [25] |
Li T, Bai G. Lesion mimic associates with adult plant resistance to leaf rust infection in wheat[J]. Theor Appl Genet, 2009, 119: 13-21.
doi: 10.1007/s00122-009-1012-7 pmid: 19330313 |
| [26] | Wang F, Wu W, Wang D, et al. Characterization and genetic analysis of a novel Light-Dependent lesion mimic mutant, lm3, showing adult-plant resistance to powdery mildew in common wheat[J]. PLos One, 2016, 11: e155358. |
| [27] |
Liang BB, Wang H, Yang C, et al. Salicylic acid is required for broad-spectrum disease resistance in rice[J]. Int J Mol Sci, 2022, 23(3): 1354.
doi: 10.3390/ijms23031354 URL |
| [28] |
Cox KL. Stronger together: Ethylene jasmonic acid, and MAPK signaling pathways synergistically induce camalexin synthesis for plant disease resistance[J]. Plant Cell, 2022, 34(8): 2829-2830.
doi: 10.1093/plcell/koac155 URL |
| [29] | Bouchez O, Huard C, Lorrain S, et al. Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1[J]. Plant Physiol, 2007, 145(2): 465-477. |
| [30] |
Wang Q, Chen X, Chai X, et al. The involvement of jasmonic acid, ethylene, and salicylic acid in the signaling pathway of clonostachys rosea-induced resistance to gray mold disease in tomato[J]. Phytopathology, 2019, 109(7): 1102-1114.
doi: 10.1094/PHYTO-01-19-0025-R URL |
| [31] |
Ding LN, Li YT, Wu YZ, et al. Plant disease resistance-related signaling pathways: recent progress and future prospects[J]. Int J Mol Sci, 2022, 23(24): 16200.
doi: 10.3390/ijms232416200 URL |
| [32] |
Yan J, Fang Y, Xue D. Advances in the genetic basis and molecular mechanism of lesion mimic formation in rice[J]. Plants, 2022, 11(16): 2169.
doi: 10.3390/plants11162169 URL |
| [33] | 王汉中. 我国油菜产业发展的历史回顾与展望[J]. 中国油料作物学报, 2010, 32(2): 300-302. |
| Wang HZ. Review and future development of rapeseed industry in China[J]. Chin J Oil Crop Sci, 2010, 32(2): 300-302. | |
| [34] |
Neik TX, Barbetti MJ, Batley J. Current status and challenges in identifying disease resistance genes in Brassica napus[J]. Front Plant Sci, 2017, 8: 1788.
doi: 10.3389/fpls.2017.01788 URL |
| [35] | Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization[J]. Japanese J Bot, 1935, 7: 389-452. |
| [36] |
Allender CJ, King GJ. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers[J]. BMC Plant Biol, 2010, 10: 54.
doi: 10.1186/1471-2229-10-54 URL |
| [37] |
Lysak MA, Koch MA, Pecinka A, et al. Chromosome triplication found across the tribe Brassiceae[J]. Genome Res, 2005, 15(4): 516-525.
doi: 10.1101/gr.3531105 pmid: 15781573 |
| [38] |
Chalhoub B, Denoeud F, Liu SY, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome[J]. Science, 2014, 345(6199): 950-953.
doi: 10.1126/science.1253435 pmid: 25146293 |
| [39] |
Beszterda M, Nogala-Kałucka M. Current research developments on the processing and improvement of the nutritional quality of rapeseed(Brassica napus L.)[J]. Eur J Lipid Sci Technol, 2019, 121(5): 1800045.
doi: 10.1002/ejlt.v121.5 URL |
| [40] | 沈金雄. 甘蓝型油菜杂种优势及其遗传分析[D]. 武汉: 华中农业大学, 2003. |
| Shen JX. Heterosis and genetic analysis of Brassica napus L.[D]. Wuhan: Huazhong Agricultural University, 2003. | |
| [41] |
Wang Y, Hou YP, Chen CJ, et al. Detection of resistance in Sclerotinia sclerotiorum to carbendazim and dimethachlon in Jiangsu Province of China[J]. Australas Plant Pathol, 2014, 43(3): 307-312.
doi: 10.1007/s13313-014-0271-1 URL |
| [42] |
Clarkson JP, Fawcett L, Anthony SG, et al. A model for Sclerotinia sclerotiorum infection and disease development in lettuce, based on the effects of temperature, relative humidity and ascospore density[J]. PLoS One, 2014, 9(4): e94049.
doi: 10.1371/journal.pone.0094049 URL |
| [43] |
Rushton PJ, Reinstädler A, Lipka V, et al. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling[J]. Plant Cell, 2002, 14(4): 749-762.
doi: 10.1105/tpc.010412 pmid: 11971132 |
| [44] |
Xie Z, Zhang ZL, Zou XL, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells[J]. Plant Physiol, 2005, 137(1): 176-189.
doi: 10.1104/pp.104.054312 pmid: 15618416 |
| [45] |
Qu D, Show PL, Miao X. Transcription factor ChbZIP1 from alkaliphilic microalgae Chlorella sp. BLD enhancing alkaline tolerance in transgenic Arabidopsis thaliana[J]. Int J Mol Sci, 2021, 22(5): 2387.
doi: 10.3390/ijms22052387 URL |
| [46] |
Buchel AS, Brederode FT, Bol JF, et al. Mutation of GT-1 binding sites in the Pr-1A promoter influences the level of inducible gene expression in vivo[J]. Plant Mol Biol, 1999, 40(3): 387-396.
pmid: 10437823 |
| [47] |
Jeong YM, Chung WH, Mun JH, et al. De novo assembly and characterization of the complete chloroplast genome of radish(Raphanus sativus L.)[J]. Gene, 2014, 551(1): 39-48.
doi: 10.1016/j.gene.2014.08.038 URL |
| [48] |
Liu SY, Liu YM, Yang XH, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes[J]. Nat Commun, 2014, 5: 3930.
doi: 10.1038/ncomms4930 |
| [1] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [2] | 蔡梦鲜, 高作敏, 胡利娟, 冯群, 王洪程, 朱斌. 天然甘蓝型油菜C染色体组C1,C2缺体的创建及遗传分析[J]. 生物技术通报, 2023, 39(3): 81-88. |
| [3] | 金姣姣, 刘自刚, 米文博, 徐明霞, 邹娅, 徐春梅, 赵彩霞. 利用RNA-Seq鉴定调控甘蓝型油菜叶片光合特性的低温胁迫应答基因[J]. 生物技术通报, 2022, 38(4): 126-142. |
| [4] | 王海波, 郭俊云. 小桐子低温诱导型启动子JcDnaJ20p的克隆及烟草转化功能鉴定[J]. 生物技术通报, 2021, 37(2): 24-31. |
| [5] | 孙威,许奕,许桂莺,孙佩光,宋顺,常胜合. 病毒诱导的基因沉默及其在植物研究中的应用[J]. 生物技术通报, 2015, 31(10): 105-110. |
| [6] | 郝晓云, 沈海涛, 李鸿彬. 甘蓝型油菜下胚轴和带柄子叶再生体系研究[J]. 生物技术通报, 2013, 0(4): 69-74. |
| [7] | 杨长友, 袁中厚, 郑小敏, 张涛. 甘蓝型油菜高效离体再生体系的建立[J]. 生物技术通报, 2013, 0(1): 111-115. |
| [8] | 孔芳;薛正莲;杨超英;王幼平;. 转Bar基因甘蓝型油菜叶片蛋白质组变化的初步分析[J]. , 2012, 0(10): 75-82. |
| [9] | 邓秋红;栗茂腾;向福;余龙江;. 甘蓝型油菜β碳酸酐酶的结构预测[J]. , 2010, 0(01): 111-117. |
| [10] | 刘建民;李运涛;甘露;李红;栗茂腾;. napin基因启动子克隆及进化分析[J]. , 2008, 0(S1): 188-191. |
| [11] | 邹智;吴刚;肖玲;武玉花;聂淑晶;肖娜;卢长明;. 植物源内含子对GUS基因表达模式的影响[J]. , 2008, 0(06): 78-82. |
| [12] | 秦春圃;. 甘蓝型油菜BnKCR2基因cDNA的克隆和真核表达载体的构建[J]. , 2008, 0(04): 201-201. |
| [13] | 路小春;李加纳;李本逊;柴友荣;. 甘蓝型油菜TT1基因反义植物表达载体的构建[J]. , 2006, 0(S1): 305-309. |
| [14] | 秦春圃. 应用RAPD标记法鉴定甘蓝型油菜杂种纯度[J]. , 2003, 0(04): 54-54. |
| [15] | 刘忠松;官春云;孟金陵;危文亮;. 油菜分子标记研究进展[J]. , 1997, 0(05): 14-16. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||