








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 128-135.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0338
侯瑞泽( ), 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰(
), 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰( )
)
收稿日期:2023-04-11
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
李梅兰,女,博士,教授,研究方向:园艺植物生物技术与遗传改良;E-mail: 15935485975@163.com作者简介:侯瑞泽,男,硕士研究生,研究方向:园艺植物生物技术与遗传改良;E-mail: hrz409972758@163.com;基金资助:
HOU Rui-ze BAO Yue CHEN Qi-liang MAO Gui-ling WEI Bo-lin HOU Lei-ping LI Mei-lan( )
)
Received:2023-04-11
Published:2023-10-26
Online:2023-11-28
摘要:
克隆BrcPRR5,并研究其在不同组织和不同发育时期的表达模式及功能,为了解PRR5对普通白菜成花转变的影响奠定基础。运用RT-PCR方法克隆PRR5的同源基因BrcPRR5,并对其进行生物信息学分析,利用荧光定量PCR测定该基因在不同组织部位和不同发育时期的相对表达量,构建过表达载体并转化拟南芥进行基因的功能验证。结果表明,BrcPRR5的CDS全长为1 701 bp,编码566个氨基酸,通过与其他物种的同源蛋白进行氨基酸序列多重比对,确定得到的序列属于普通白菜PRR5同源基因。BrcPRR5在茎和花中的表达量高于叶和果荚中的表达量;在S0茎尖中表达量最高,说明其在普通白菜成花转变过程中表达量上调可能促进成花转变。超表达转基因植株开花期提前,株高和茎粗明显优于野生型。BrcPRR5可以促进植株提早抽薹开花。
侯瑞泽, 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰. 普通白菜PRR5的克隆、表达及功能验证[J]. 生物技术通报, 2023, 39(10): 128-135.
HOU Rui-ze BAO Yue CHEN Qi-liang MAO Gui-ling WEI Bo-lin HOU Lei-ping LI Mei-lan. Cloning,Expression and Functional Identification of PRR5 Gene in Pakchoi[J]. Biotechnology Bulletin, 2023, 39(10): 128-135.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer purpose |
|---|---|---|
| C-Bra009768 | F:ATGGGAGAAGTGAGCGACGAAG | 基因克隆 |
| R:TCATTGTGGAGCTTCTTGTGTTGAG | Gene clone | |
| G-Bra009768 | F:GAGAACACGGGGGACTCTAGAATGGGAGAAGTGAGCGACGAAG | 基因克隆 Gene clone |
| R:ATAAGGGACTGACCACCCGGGTCATTGTGGAGCTTCTTGTGTTGAG | ||
| N-YZ | F:GACGTTCCAACCACGTCTTC | 菌液PCR引物 |
| R:CCAGACTGAATGCCCACAGG | Bacterial liquid PCR primer |
表1 Bra009768引物列表
Table 1 Primers of Bra009768
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer purpose |
|---|---|---|
| C-Bra009768 | F:ATGGGAGAAGTGAGCGACGAAG | 基因克隆 |
| R:TCATTGTGGAGCTTCTTGTGTTGAG | Gene clone | |
| G-Bra009768 | F:GAGAACACGGGGGACTCTAGAATGGGAGAAGTGAGCGACGAAG | 基因克隆 Gene clone |
| R:ATAAGGGACTGACCACCCGGGTCATTGTGGAGCTTCTTGTGTTGAG | ||
| N-YZ | F:GACGTTCCAACCACGTCTTC | 菌液PCR引物 |
| R:CCAGACTGAATGCCCACAGG | Bacterial liquid PCR primer |
| 引物名称 Gene name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer purpose |
|---|---|---|
| ACTIN | F:GTTGCTATCCAGGCTGTTCT | 大白菜内参引物 |
| R:AGCGTGAGGAAGAGCATAAC | Internal primers in Brassica pekinensis | |
| Bra009768 | F:AGTTACAGAGTCGCTGCT | 荧光定量引物 |
| R:TATTAACCGAGTCCTGTGTTG | Fluorescent quantitative primer | |
| ACT11 | F:CACACTGGAGTGATGGTTGG | 拟南芥内参引物 |
| R:ATTGGCCTTGGGGTTAAGAG | The internal reference gene of Arabidopsis |
表2 RT-qPCR验证引物
Table 2 Primers of real time quantitative PCR
| 引物名称 Gene name | 引物序列 Primer sequence(5'-3') | 引物用途 Primer purpose |
|---|---|---|
| ACTIN | F:GTTGCTATCCAGGCTGTTCT | 大白菜内参引物 |
| R:AGCGTGAGGAAGAGCATAAC | Internal primers in Brassica pekinensis | |
| Bra009768 | F:AGTTACAGAGTCGCTGCT | 荧光定量引物 |
| R:TATTAACCGAGTCCTGTGTTG | Fluorescent quantitative primer | |
| ACT11 | F:CACACTGGAGTGATGGTTGG | 拟南芥内参引物 |
| R:ATTGGCCTTGGGGTTAAGAG | The internal reference gene of Arabidopsis |
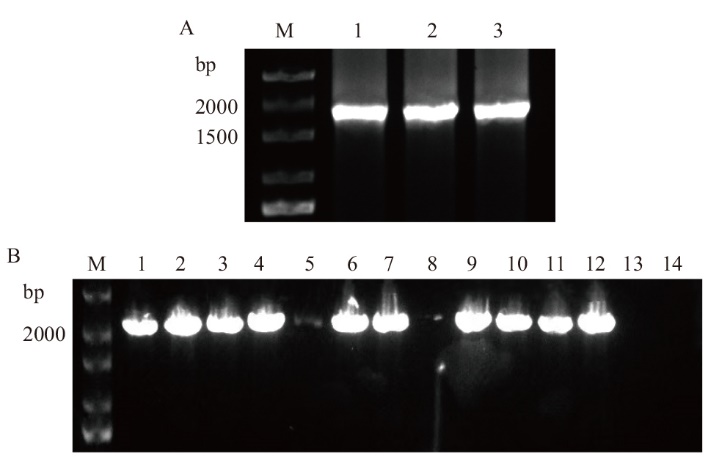
图1 Bra009768的克隆及其菌液检测结果 A:Bra009768的克隆(M:DNA marker;1-3:Bra009768);B:菌液的PCR检测(M:DNA maker;1-12:Bra009768;13-14:阴性对照)
Fig. 1 Cloning of Bra009768 and detection results of its broth A: Cloning of Bra009768(M: DNA marker; 1-3: Bra009768). B: PCR detection of bacterial liquid(M: DNA marker; 1-12: Bra009768; 13-14: negative control)
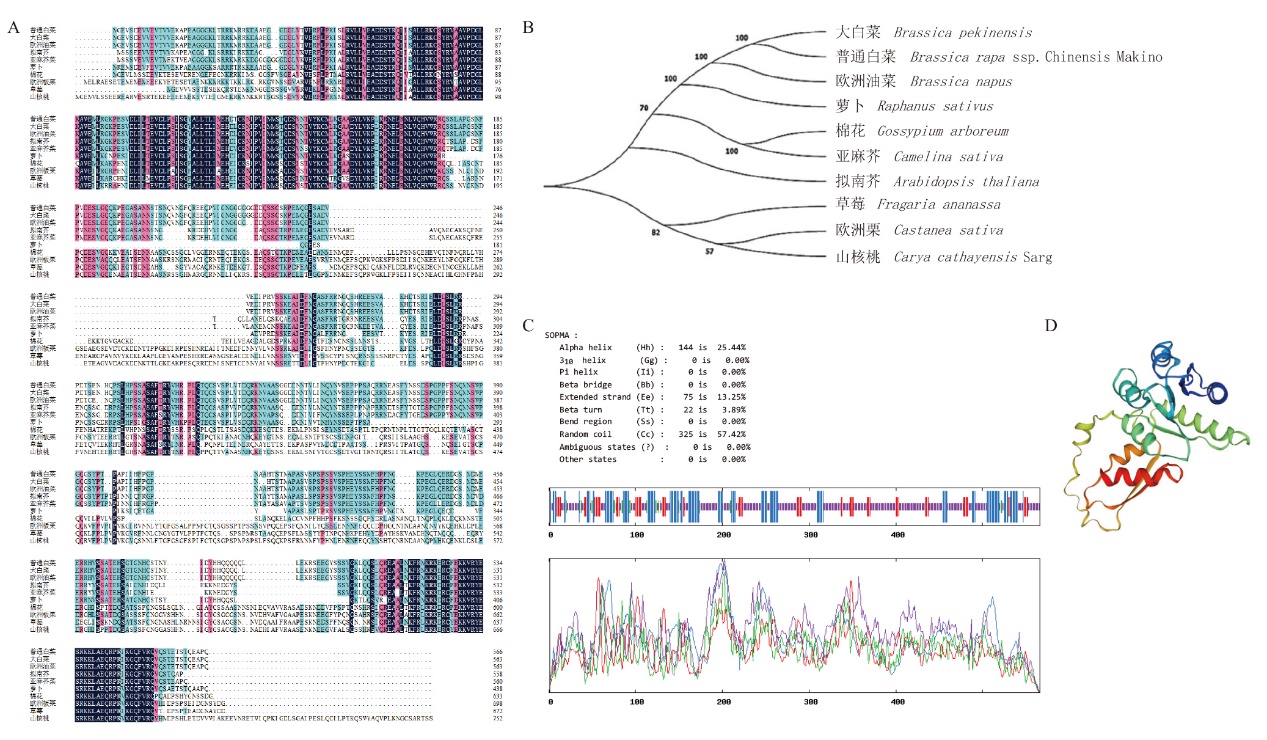
图2 Bra009768的生物信息学分析A:Bra009768氨基酸序列多重比对;B:Bra009768同源蛋白的系统进化树;C、D:Bra009768蛋白质二级、三级结构预测
Fig. 2 Bioinformatics analysis of Bra009768 A: Multiple alignment of Bra009768 amino acid sequences. B: Phylogenetic tree of Bra009768 homologous proteins. C, D: Prediction of secondary and tertiary structures of Bra009768 proteins
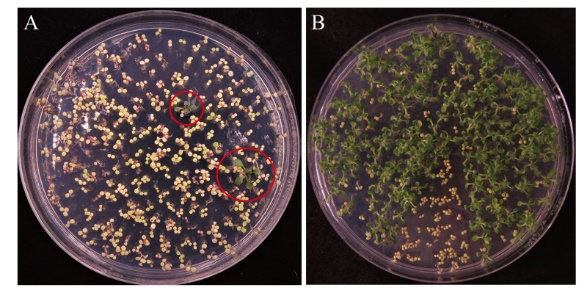
图4 转基因拟南芥抗性植株筛选 A:T1代抗性植株筛选;B:T2代抗性植株筛选
Fig. 4 Selection of transgenic Arabidopsis thaliana resistant plants A: Screening of resistant plants in T1 generation. B: Screening of resistant plants in T2 generation
| 植株 Plant | 抽薹时间 Bolting time/d | 侧薹数 Number of side bolting | 株高 Plant height/cm | 茎粗 Stem thick/mm |
|---|---|---|---|---|
| Col-1 | 13.75±0.82 | 11.50±0.31 | 25.90±0.88 | 0.45±0.04 |
| T-1 | 12.50±2.38 | 11.25±2.12 | 25.75±3.13 | 0.54±0.12 |
| Col-2 | 17.20±1.25 | 7.50±0.21 | 22.30±0.91 | 0.53±0.02 |
| T-2 | 15.72±0.78 | 8.10±0.13 | 29.80±0.53 | 0.94±0.06 |
表3 T1、T2转基因植株表型统计
Table 3 Phenotypic statistics of T1 and T2 transgenic plants
| 植株 Plant | 抽薹时间 Bolting time/d | 侧薹数 Number of side bolting | 株高 Plant height/cm | 茎粗 Stem thick/mm |
|---|---|---|---|---|
| Col-1 | 13.75±0.82 | 11.50±0.31 | 25.90±0.88 | 0.45±0.04 |
| T-1 | 12.50±2.38 | 11.25±2.12 | 25.75±3.13 | 0.54±0.12 |
| Col-2 | 17.20±1.25 | 7.50±0.21 | 22.30±0.91 | 0.53±0.02 |
| T-2 | 15.72±0.78 | 8.10±0.13 | 29.80±0.53 | 0.94±0.06 |
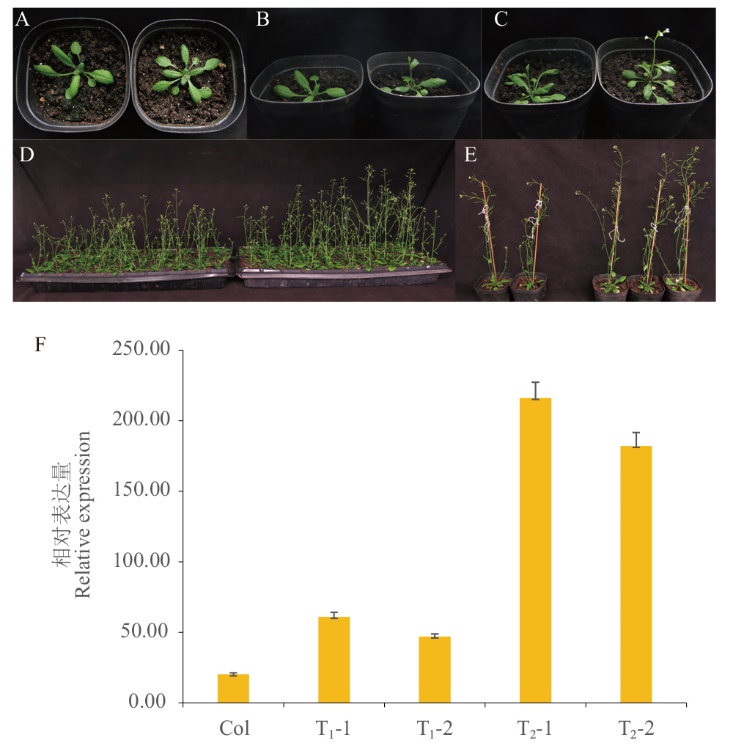
图5 转基因植株的表型观察及BrcPRR5的RT-qPCR验证 A:营养生长期;B:抽薹期;C:现蕾期;D-E:成熟期(左:野生型(Col);右:转基因植株);F:转基因植株的RT-qPCR检测
Fig. 5 Phenotypic observation of transgenic plants and RT-qPCR validation of BrcPRR5 A: Vegetative growth period. B: Bolting stage. C: Budding stage. D-E: Mature stage(left: wild type(Col); right: transgenic plants). F: RT-qPCR detection of transgenic plants
| [25] |
Imaizumi T, Tran HG, Swartz TE, et al. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis[J]. Nature, 2003, 426(6964): 302-306.
doi: 10.1038/nature02090 |
| [26] |
Imaizumi T, Schultz TF, Harmon FG, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis[J]. Science, 2005, 309(5732): 293-297.
doi: 10.1126/science.1110586 pmid: 16002617 |
| [27] |
Fornara F, Panigrahi KCS, Gissot L, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response[J]. Dev Cell, 2009, 17(1): 75-86.
doi: 10.1016/j.devcel.2009.06.015 pmid: 19619493 |
| [28] |
Valverde F, Mouradov A, Soppe W, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering[J]. Science, 2004, 303(5660): 1003-1006.
doi: 10.1126/science.1091761 pmid: 14963328 |
| [29] |
Nakamichi N, Kita M, Niinuma K, et al. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway[J]. Plant Cell Physiol, 2007, 48(6): 822-832.
doi: 10.1093/pcp/pcm056 pmid: 17504813 |
| [30] |
Hayama R, Sarid-Krebs L, Richter R, et al. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length[J]. Embo J, 2017, 36(7): 904-918.
doi: 10.15252/embj.201693907 pmid: 28270524 |
| [1] |
Amasino R. Seasonal and developmental timing of flowering[J]. Plant J, 2010, 61(6): 1001-1013.
doi: 10.1111/tpj.2010.61.issue-6 URL |
| [2] |
Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis[J]. Cell, 2010, 141(3): 550-550.e2.
doi: 10.1016/j.cell.2010.04.024 pmid: 20434991 |
| [3] | 刘娟, 黎黎, 陆柄辰, 等. 温度调控植物开花研究进展[J]. 应用与环境生物学报, 2020, 26(3): 713-721. |
| Liu J, Li L, Lu BC, et al. Research progress on temperature regulation of plant flowering[J]. Chin J Appl Environ Biol, 2020, 26(3): 713-721. | |
| [4] | 孙昌辉, 邓晓建, 方军, 等. 高等植物开花诱导研究进展[J]. 遗传, 2007, 29(10): 1182-1190. |
| Sun CH, Deng XJ, Fang J, et al. An overview of flowering transition in higher plants[J]. Hereditas, 2007, 29(10): 1182-1190. | |
| [5] |
Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues[J]. Nat rev Genet, 2012, 13(9): 627-639.
doi: 10.1038/nrg3291 pmid: 22898651 |
| [6] |
Mutasa-Göttgens E, Hedden P. Gibberellin as a factor in floral regulatory networks[J]. J Exp Bot, 2009, 60(7): 1979-1989.
doi: 10.1093/jxb/erp040 pmid: 19264752 |
| [7] |
Bäurle I, Dean C. The timing of developmental transitions in plants[J]. Cell, 2006, 125(4): 655-664.
doi: 10.1016/j.cell.2006.05.005 pmid: 16713560 |
| [8] |
Sablowski R. Flowering and determinacy in Arabidopsis[J]. J Exp Bot, 2007, 58(5): 899-907.
doi: 10.1093/jxb/erm002 pmid: 17293602 |
| [9] | Boss PK, Bastow RM, Mylne JS, et al. Multiple pathways in the decision to flower: Enabling, promoting, and resetting[J]. Plant Cell, 2004, 16(suppl): S18-S31. |
| [10] |
Makino S, Kiba T, Imamura A, et al. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana[J]. Plant Cell Physiol, 2000, 41(6): 791-803.
doi: 10.1093/pcp/41.6.791 pmid: 10945350 |
| [11] |
Nakamichi N, Kiba T, Kamioka M, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways[J]. Proc Natl Acad Sci USA, 2012, 109(42): 17123-17128.
doi: 10.1073/pnas.1205156109 pmid: 23027938 |
| [12] |
Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence[J]. Trends Plant Sci, 2006, 11(11): 550-558.
pmid: 17035069 |
| [13] |
Nakamichi N, Kudo T, Makita N, et al. Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5[J]. Biosci, Biotechnol, Biochem, 2020, 84(5): 970-979.
doi: 10.1080/09168451.2020.1719822 URL |
| [14] |
Shang M, Wang X, Zhang J, et al. Genetic regulation of GA metabolism during vernalization, floral bud initiation and development in pak choi(Brassica rapa ssp. chinensis Makino)[J]. Front Plant Sci, 2017, 8: 1533.
doi: 10.3389/fpls.2017.01533 URL |
| [15] | 宋红霞, 付超, 侯雷平, 等. 普通白菜石蜡切片染色方法筛选及花芽分化形态学鉴定[J]. 江苏农业科学, 2018, 46(2): 88-91. |
| Song HX, Fu C, Hou LP, et al. Screening of staining methods for paraffin sections of Chinese cabbage and morphological identification of flower bud differentiation[J]. Jiangsu Agric Sci, 2018, 46(2): 88-91. | |
| [16] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT)method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [17] |
Strayer C, Oyama T, Schultz TF, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog[J]. Science, 2000, 289(5480): 768-771.
doi: 10.1126/science.289.5480.768 pmid: 10926537 |
| [18] |
Ito S, Matsushika A, Yamada H, et al. Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana[J]. Plant Cell Physiol, 2003, 44(11): 1237-1245.
doi: 10.1093/pcp/pcg136 URL |
| [19] |
Yamamoto Y, Sato E, Shimizu T, et al. Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis[J]. Plant Cell Physiol, 2003, 44(11): 1119-1130.
pmid: 14634148 |
| [20] |
Murakami M, Yamashino T, Mizuno T. Characterization of circadian-associated APRR3 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana[J]. Plant Cell Physiol, 2004, 45(5): 645-650.
pmid: 15169947 |
| [21] |
Para A, Farré EM, Imaizumi T, et al. PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock[J]. Plant Cell, 2007, 19(11): 3462-3473.
doi: 10.1105/tpc.107.054775 URL |
| [22] |
Nakamichi N, Takao SR, Kudo T, et al. Improvement of Arabidopsis biomass and cold, drought and salinity stress tolerance by modified circadian clock-associated PSEUDO-RESPONSE REGULATORs[J]. Plant Cell Physiol, 2016, 57(5): 1085-1097.
doi: 10.1093/pcp/pcw057 pmid: 27012548 |
| [23] |
Matsushika A, Murakami M, Ito S, et al. Characterization of Circadian-associated pseudo-response regulators: I. Comparative studies on a series of transgenic lines misexpressing five distinctive PRR Genes in Arabidopsis thaliana[J]. Biosci, Biotechnol, Biochem, 2007, 71(2): 527-534.
doi: 10.1271/bbb.60583 URL |
| [24] |
Beales J, Turner A, Griffiths S, et al. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat(Triticum aestivum L.)[J]. Theor Appl Genet, 2007, 115(5): 721-733.
doi: 10.1007/s00122-007-0603-4 pmid: 17634915 |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [4] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [5] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [6] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [7] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| [8] | 王楠, 张瑞, 潘阳阳, 何翃宏, 王靖雷, 崔燕, 余四九. 牦牛TGF-β1基因克隆及在雌性生殖系统主要器官中的表达定位[J]. 生物技术通报, 2022, 38(6): 279-290. |
| [9] | 李洋, 张晓天, 朴静子, 周如军, 李自博, 关海雯. 花生疮痂病菌蓝光受体EaWC 1基因克隆及生物信息学分析[J]. 生物技术通报, 2022, 38(5): 93-99. |
| [10] | 胡琪, 侯玉翔, 李璿, 李梅兰. 普通白菜CYP79B2同源基因的克隆与表达[J]. 生物技术通报, 2022, 38(12): 168-174. |
| [11] | 甘诚燕, 张心慧, 王沙, 樊瑶羽薇, 招雪晴, 苑兆和. 石榴花发育相关基因PgSPL2的克隆及功能研究[J]. 生物技术通报, 2022, 38(12): 194-203. |
| [12] | 盛雪晴, 赵楠, 林亚秋, 陈定双, 王瑞龙, 李傲, 王永, 李艳艳. 山羊ZNF32的克隆及表达分析[J]. 生物技术通报, 2022, 38(12): 300-311. |
| [13] | 付伟杰, 邝杰华, 罗君, 黄建盛, 陈有铭, 陈刚. 杉虎斑Galectin-8基因克隆及其在不同阿魏酸水平饲料下的表达响应[J]. 生物技术通报, 2022, 38(12): 312-323. |
| [14] | 张琳, 魏祯祯, 宋程威, 郭丽丽, 郭琪, 侯小改, 王华芳. ‘凤丹’牡丹PoFD基因克隆及表达分析[J]. 生物技术通报, 2022, 38(11): 104-111. |
| [15] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||