








生物技术通报 ›› 2023, Vol. 39 ›› Issue (7): 151-159.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1408
收稿日期:2022-11-15
出版日期:2023-07-26
发布日期:2023-08-17
通讯作者:
王文杰,男,研究员,研究方向:茶树品种、茶叶加工与品质;E-mail: 391590137@qq.com作者简介:孙明慧,女,研究实习员,研究方向:茶树种质资源与遗传育种;E-mail: sunminghui@aaas.org.cn
基金资助:
SUN Ming-hui( ), WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie(
), WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie( )
)
Received:2022-11-15
Published:2023-07-26
Online:2023-08-17
摘要:
基于茶树全基因组数据,筛选鉴定茶树(Camellia sinensis(L.)O. Ktze.)TATA元件调控因子(TATA element modulatory factor,TMF)CsTMFs家族成员。克隆茶树叶片中CsTMFs编码区序列(coding sequence,CDS)全长。使用生物信息学方法对CsTMFs的保守结构域、基因结构、理化性质、蛋白质二级结构和系统发育关系进行分析。基于转录组数据进行组织特异性表达的分析,利用实时荧光定量聚合酶链式反应(real-time fluorescent quantitative PCR,qPCR)方法检测冷胁迫和干旱胁迫下CsTMFs在茶树叶片中的表达情况。结果表明,CsTMFs家族有2个CsTMF基因,序列CDS区长度分别为2 934和2 904 bp,均具有TMF-DNA-bd和TMF-TATA-bd完整保守结构域。CsTMF1和CsTMF2具有组织表达特异性,CsTMF1在花与茎中表达较高,CsTMF2在成熟叶与茎中表达较高。CsTMF1受冷胁迫诱导,CsTMF2受冷胁迫和干旱胁迫抑制。
孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159.
SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant[J]. Biotechnology Bulletin, 2023, 39(7): 151-159.
| 用途Application | 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| 实时荧光定量PCR qPCR | CsTMF1-qF | TCCATCCGTGACTCTCTTGCTGAG |
| CsTMF1-qR | GCCTCCGCCTTAGTGCCTCTA | |
| CsTMF2-qF | CGACTGGCATCTCTGGAATCC | |
| CsTMF2-qR | GCCTCCGTCTTAATGCTTCTAACT | |
| Actin-qF | GCCATCTTTGATTGGAATGG | |
| Actin-qR | GGTGCCACAACCTTGATCTT | |
| 基因克隆Gene cloning | CsTMF1-F | ATGGCCTGGTTTAGCGGGAAAGTCTCTTTGG |
| CsTMF1-R | TCATGCAGAGCCCATAGAAGAACCCATTACC | |
| CsTMF2-F | ATGGCCTGGTTCGGTGGGAAAGTCT | |
| CsTMF2-R | CTAAGACGAACTTAGTACCTGGATC |
表1 PCR 引物信息
Table 1 Primer sequences for PCR
| 用途Application | 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|---|
| 实时荧光定量PCR qPCR | CsTMF1-qF | TCCATCCGTGACTCTCTTGCTGAG |
| CsTMF1-qR | GCCTCCGCCTTAGTGCCTCTA | |
| CsTMF2-qF | CGACTGGCATCTCTGGAATCC | |
| CsTMF2-qR | GCCTCCGTCTTAATGCTTCTAACT | |
| Actin-qF | GCCATCTTTGATTGGAATGG | |
| Actin-qR | GGTGCCACAACCTTGATCTT | |
| 基因克隆Gene cloning | CsTMF1-F | ATGGCCTGGTTTAGCGGGAAAGTCTCTTTGG |
| CsTMF1-R | TCATGCAGAGCCCATAGAAGAACCCATTACC | |
| CsTMF2-F | ATGGCCTGGTTCGGTGGGAAAGTCT | |
| CsTMF2-R | CTAAGACGAACTTAGTACCTGGATC |
| Motif | 宽度Width | 最可能序列Most possible match | 结构域Domain |
|---|---|---|---|
| 1 | 80 | TARLQPSTVPDQSPITRRKSGFENGNLSRKLSSASSLGSMEESFFLQASLDSSDNFSERRNPGEMAMSPYFMKSMPPSAF | * |
| 2 | 78 | TMLVQALEELRQTLSRKEQQAVFREDMLRRDIEDLQKRYQESERRCEELVTQVPESTRPLLRQIEAMQETTGRRAEAW | * |
| 3 | 40 | MAWFGGKVSLGNFPDLAGAVNKISESVKNIEKNFDSALGF | * |
| 4 | 80 | MKMMETALQGAARQAQAKADEIAKLMNENENQKAVIEDLRRKSNEAEIESLREEYHQKVAALERKVYALTKERDTLRREQ | * |
| 5 | 80 | IRDSLAEELVKMTAQCEKLRVEAAILPGIRAELEALRRRHSAALELMGERDEELEELRADIVDLKEMYREQVNLLVNKIQ | TMF-TATA-bd |
| 6 | 79 | GLWPSGTDRKTJFEPVIAFMGHKGGESTVESIEKPELLEPPSSVEEKZEGENDRSTDFATEQTRPAEEVNZESSHMLPD | * |
| 7 | 79 | QAGMEVELPGFGFPTSKEIESAEEHFTDDLPDGPPPDEAAEMVSEPVSHENDTFGREVELNZQASDYEPDIKEQRVSSG | TMF-DNA-bd |
| 8 | 80 | VMAEGEELSKKQAAQESQIRKLRAQIRELEEEKKGLTTKLQVEENKVESIKRDKAETEKLLQETIEKHQAEJAVQKEFYT | * |
| 9 | 80 | QTLSRLNVLEAQISCLRAEQTQLSRSLEKERQRAAENRQEYLAAKEEADTHEGRVSQLEEEIRELRRKQKZELQEALMHR | * |
| 10 | 66 | TSALGPFENSELVEAKPGTNEVDQVEVGAPVPPESHNVINLHEISDEQKAZEEEIVENLSPVQAED | * |
表2 CsTMFs蛋白序列中10个保守基序的详细信息
Table 2 Detailed information of 10 motifs in the CsTMFs proteins
| Motif | 宽度Width | 最可能序列Most possible match | 结构域Domain |
|---|---|---|---|
| 1 | 80 | TARLQPSTVPDQSPITRRKSGFENGNLSRKLSSASSLGSMEESFFLQASLDSSDNFSERRNPGEMAMSPYFMKSMPPSAF | * |
| 2 | 78 | TMLVQALEELRQTLSRKEQQAVFREDMLRRDIEDLQKRYQESERRCEELVTQVPESTRPLLRQIEAMQETTGRRAEAW | * |
| 3 | 40 | MAWFGGKVSLGNFPDLAGAVNKISESVKNIEKNFDSALGF | * |
| 4 | 80 | MKMMETALQGAARQAQAKADEIAKLMNENENQKAVIEDLRRKSNEAEIESLREEYHQKVAALERKVYALTKERDTLRREQ | * |
| 5 | 80 | IRDSLAEELVKMTAQCEKLRVEAAILPGIRAELEALRRRHSAALELMGERDEELEELRADIVDLKEMYREQVNLLVNKIQ | TMF-TATA-bd |
| 6 | 79 | GLWPSGTDRKTJFEPVIAFMGHKGGESTVESIEKPELLEPPSSVEEKZEGENDRSTDFATEQTRPAEEVNZESSHMLPD | * |
| 7 | 79 | QAGMEVELPGFGFPTSKEIESAEEHFTDDLPDGPPPDEAAEMVSEPVSHENDTFGREVELNZQASDYEPDIKEQRVSSG | TMF-DNA-bd |
| 8 | 80 | VMAEGEELSKKQAAQESQIRKLRAQIRELEEEKKGLTTKLQVEENKVESIKRDKAETEKLLQETIEKHQAEJAVQKEFYT | * |
| 9 | 80 | QTLSRLNVLEAQISCLRAEQTQLSRSLEKERQRAAENRQEYLAAKEEADTHEGRVSQLEEEIRELRRKQKZELQEALMHR | * |
| 10 | 66 | TSALGPFENSELVEAKPGTNEVDQVEVGAPVPPESHNVINLHEISDEQKAZEEEIVENLSPVQAED | * |
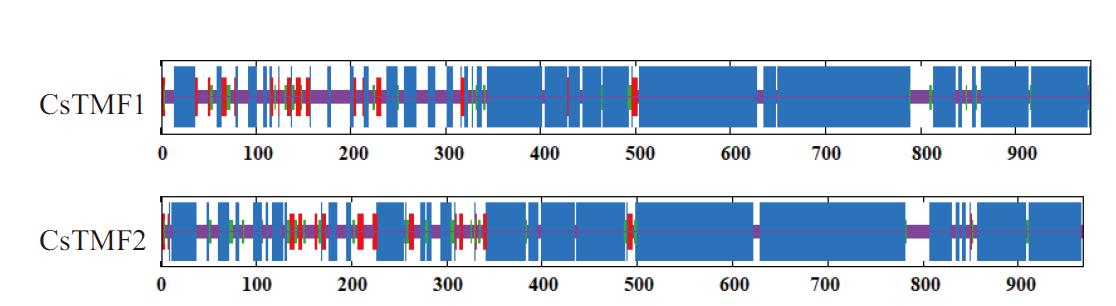
图4 CsTMFs蛋白二级结构预测 α-螺旋(蓝色)、延伸链(红色)、β-转角(绿色)和随机线圈(紫色)
Fig. 4 Secondary structure prediction of CsTMFs protein α-helix(blue), extended strand(red), β-turn(green)and random coil(purple)
| 物种名Species name | 登录号Accession No. |
|---|---|
| 水稻Oryza sativa Japonica Group | BAG90311.1 |
| 茭白Zizania palustris | KAG8086397 |
| 小麦Triticum aestivum | XP 044346450.1 |
| 大麦Hordeum vulgare subsp. Vulgar | BAJ99499 |
| 玉米Zea mays | XP 008656031.1 |
表3 不同物种TMF登录号
Table 3 Accession numbers of different species of TMF
| 物种名Species name | 登录号Accession No. |
|---|---|
| 水稻Oryza sativa Japonica Group | BAG90311.1 |
| 茭白Zizania palustris | KAG8086397 |
| 小麦Triticum aestivum | XP 044346450.1 |
| 大麦Hordeum vulgare subsp. Vulgar | BAJ99499 |
| 玉米Zea mays | XP 008656031.1 |
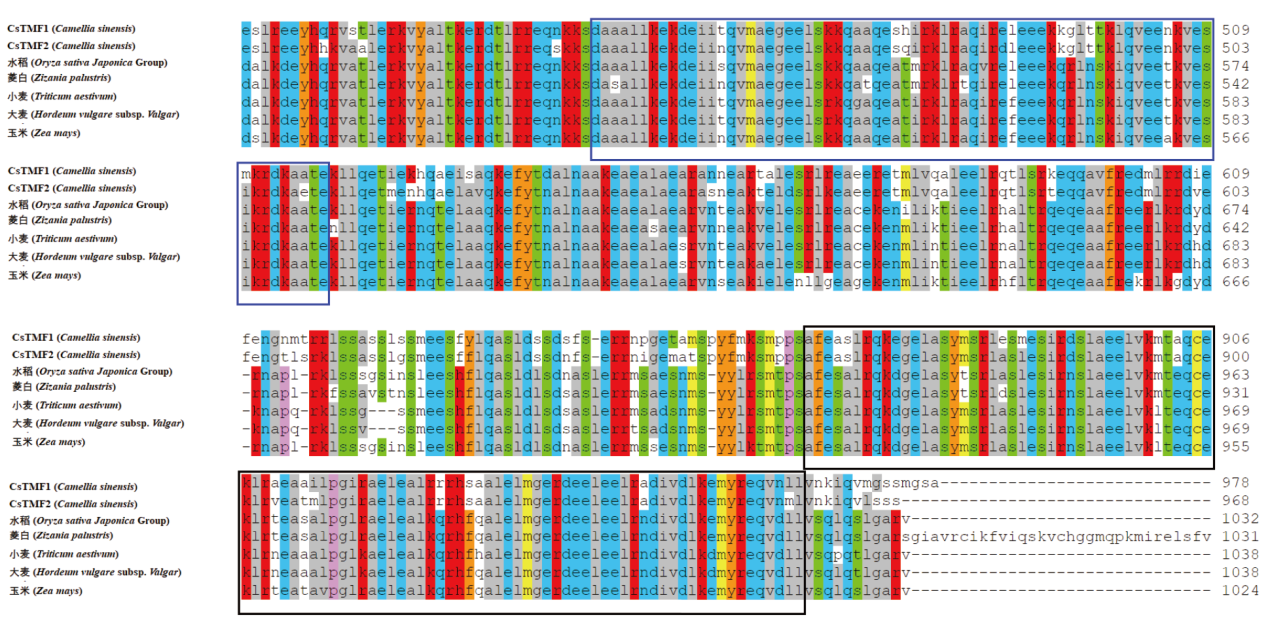
图8 茶树与其他植物TMF的多重序列比对 紫色框内为TMF-DNA-bd保守结构域,黑色框内为TMF-TATA-bd保守结构域
Fig. 8 Multiple sequence alignment of tea plant TMF with other plants The conserved domain of TMF-DNA-bd is in purple box, and the conserved domain of TMF-TATA-bd is in black box
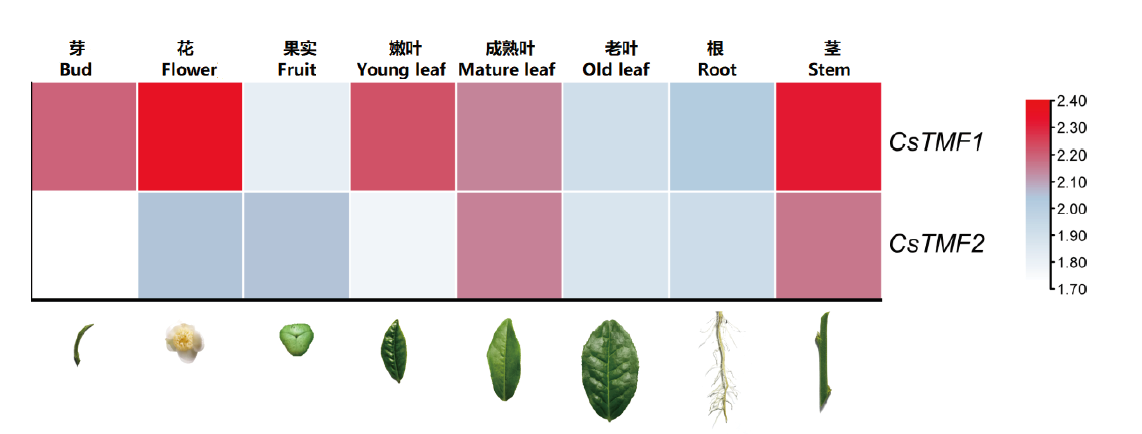
图9 CsTMF1与CsTMF2在茶树不同组织中的表达情况 热图体现的log5(FPKM+1)值
Fig. 9 Expressions of CsTMF1 and CsTMF2 in different tissues of tea plant The log5(FPKM+1)value reflects in the heatmap

图10 CsTMFs在低温和干旱胁迫下的表达模式 不同小写字母代表同一种处理不同时间点差异显著(P<0.05)
Fig. 10 Expression pattern of CsTMFs gene under drought stress and low temperature Different lowercase letters in figure indicate significant differences at different time of the same treatment(P<0.05)
| [1] | 徐燕, 朱创, 邰玲玲, 等. 红茶化学成分及生理活性的研究进展[J]. 安徽农业大学学报, 2020, 47(5): 687-696. |
| Xu Y, Zhu C, Tai LL, et al. Research progress on chemical constituents and physiological activities of black tea[J]. J Anhui Agric Univ, 2020, 47(5): 687-696. | |
| [2] | 陈思文, 康芮, 郭志远, 等. 茶树CsCML16基因的克隆及其低温胁迫下的表达分析[J]. 茶叶科学, 2021, 41(3): 315-326. |
| Chen SW, Kang R, Guo ZY, et al. Cloning and expression analysis of CsCML16 in tea plants(Camellia sinensis)under low temperature stress[J]. J Tea Sci, 2021, 41(3): 315-326. | |
| [3] | 刘声传, 陈亮. 茶树耐旱机理及抗旱节水研究进展[J]. 茶叶科学, 2014, 34(2): 111-121. |
| Liu SC, Chen L. Research advances on the drought-resistance mechanism and strategy of tea plant[J]. J Tea Sci, 2014, 34(2): 111-121. | |
| [4] | Zhou CZ, Zhu C, Fu HF, et al. Genome-wide investigation of superoxide dismutase(SOD)gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant(Camellia sinensis)[J]. PLoS One, 2019, 14(10): e0223609. |
| [5] |
Garcia JA, Ou SH, Wu F, et al. Cloning and chromosomal mapping of a human immunodeficiency virus 1 TATA element modulatory factor[J]. Proc Natl Acad Sci USA, 1992, 89(20): 9372-9376.
pmid: 1409643 |
| [6] |
Elkis Y, Bel S, Lerer-Goldstein T, et al. Testosterone deficiency accompanied by testicular and epididymal abnormalities in TMF-/- mice[J]. Mol Cell Endocrinol, 2013, 365(1): 52-63.
doi: 10.1016/j.mce.2012.09.003 pmid: 23000399 |
| [7] |
Fridmann-Sirkis Y, Siniossoglou S, Pelham HRB. TMF is a golgin that binds Rab6 and influences Golgi morphology[J]. BMC Cell Biol, 2004, 5: 18.
pmid: 15128430 |
| [8] |
Abrham G, Volpe M, Shpungin S, et al. TMF/ARA160 downregulates proangiogenic genes and attenuates the progression of PC3 xenografts[J]. Int J Cancer, 2009, 125(1): 43-53.
doi: 10.1002/ijc.24277 pmid: 19330832 |
| [9] | 徐艳. 水稻OsSKIPa互作蛋白OsTMF和OsARID3在抗逆和发育中的功能研究[D]. 武汉: 华中农业大学, 2015. |
| Xu Y. Study on the function of rice OsSKIPa interaction proteins OsTMF and OsARID3 in stress resistance and development[D]. Wuhan: Huazhong Agricultural University, 2015. | |
| [10] |
Xu Y, Hu D, Hou X, et al. OsTMF attenuates cold tolerance by affecting cell wall properties in rice[J]. New Phytol, 2020, 227(2): 498-512.
doi: 10.1111/nph.16549 pmid: 32176820 |
| [11] |
Miura K, Furumoto T. Cold signaling and cold response in plants[J]. Int J Mol Sci, 2013, 14(3): 5312-5337.
doi: 10.3390/ijms14035312 pmid: 23466881 |
| [12] |
Chinnusamy V, Zhu JH, Zhu JK. Cold stress regulation of gene expression in plants[J]. Trends Plant Sci, 2007, 12(10): 444-451.
doi: 10.1016/j.tplants.2007.07.002 pmid: 17855156 |
| [13] |
Latijnhouwers M, Gillespie T, Boevink P, et al. Localization and domain characterization of Arabidopsis golgin candidates[J]. J Exp Bot, 2007, 58(15/16): 4373-4386.
doi: 10.1093/jxb/erm304 URL |
| [14] |
Li YY, Wang XW, Ban QY, et al. Comparative transcriptomic analysis reveals gene expression associated with cold adaptation in the tea plant Camellia sinensis[J]. BMC Genomics, 2019, 20(1): 624.
doi: 10.1186/s12864-019-5988-3 |
| [15] | 唐磊, 肖罗丹, 黄伊凡, 等. 茶树CsMGTs基因的克隆及其镁转运功能分析[J]. 茶叶科学, 2021, 41(6): 761-776. |
| Tang L, Xiao LD, Huang YF, et al. Cloning of CsMGTs gene from tea plant and analysis of its magnesium transport function[J]. J Tea Sci, 2021, 41(6): 761-776. | |
| [16] |
Wu Q, Tong W, Zhao HJ, et al. Comparative transcriptomic analysis unveils the deep phylogeny and secondary metabolite evolution of 116 Camellia plants[J]. Plant J, 2022, 111(2): 406-421.
doi: 10.1111/tpj.v111.2 URL |
| [17] |
Li P, Guo WZ. Genome-wide characterization of the Rab gene family in Gossypium by comparative analysis[J]. Bot Stud, 2017, 58(1): 26.
doi: 10.1186/s40529-017-0181-y URL |
| [18] | 裴惠娟, 张满效, 安黎哲. 非生物胁迫下植物细胞壁组分变化[J]. 生态学杂志, 2011, 30(6): 1279-1286. |
| Pei HJ, Zhang MX, An LZ. Changes of plant cell wall components under abiotic stresses: a review[J]. Chin J Ecol, 2011, 30(6): 1279-1286. | |
| [19] | 陈媛媛. 利用CRISPR/Cas9技术初步解析水稻纤维素合酶(OsCESA4和OsCESA7)P-CR区的功能[D]. 武汉: 华中农业大学, 2018. |
| Chen YY.The function of P-Cr region of rice cellulose synthase(OsCESA4 and OSCESA7)was preliminarily analyzed by CRISPR/Cas 9 technology.[D]. Wuhan: Huazhong Agricultural University, 2018. | |
| [20] |
Liu HH, Ma Y, Chen N, et al. Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice[J]. Plant Cell Environ, 2014, 37(5): 1144-1158..
doi: 10.1111/pce.2014.37.issue-5 URL |
| [1] | 赵光绪, 杨合同, 邵晓波, 崔志豪, 刘红光, 张杰. 一株高效溶磷产红青霉培养条件优化及其溶磷特性[J]. 生物技术通报, 2023, 39(9): 71-83. |
| [2] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [3] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [4] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [5] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [6] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [7] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [8] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [9] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [10] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [11] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [12] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [13] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [14] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [15] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||