








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 143-151.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0824
肖小军1,2( ), 陈明2, 韩德鹏2, 余跑兰2, 郑伟2, 肖国滨2, 周庆红1(
), 陈明2, 韩德鹏2, 余跑兰2, 郑伟2, 肖国滨2, 周庆红1( ), 周会汶3(
), 周会汶3( )
)
收稿日期:2022-07-04
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
周庆红,女,教授,研究方向:作物育种;E-mail:qinghongzhou@126.com;作者简介:肖小军,男,博士研究生,研究方向:油菜遗传育种;E-mail:xiao850908@163.com基金资助:
XIAO Xiao-jun1,2( ), CHEN Ming2, HAN De-peng2, YU Pao-lan2, ZHENG Wei2, XIAO Guo-bin2, ZHOU Qing-hong1(
), CHEN Ming2, HAN De-peng2, YU Pao-lan2, ZHENG Wei2, XIAO Guo-bin2, ZHOU Qing-hong1( ), ZHOU Hui-wen3(
), ZHOU Hui-wen3( )
)
Received:2022-07-04
Published:2023-03-26
Online:2023-04-10
摘要:
为挖掘甘蓝型油菜每角果粒数显著关联单核苷酸多态性(SNP)位点及相关候选基因。本研究以300份甘蓝型油菜自交系为试验材料,对甘蓝油菜每角果粒数进行一年两地表型考察,并结合该群体前期开发的201 817个SNPs标记,采用一般线性模型(GLM)和混合线性模型(MLM)进行全基因组关联分析(GWAS),此外,对性状显著关联SNP位点两侧100 kb区域内相关候选基因进行功能预测。300份甘蓝型油菜每角果粒数在两地均表现出广泛的表型变异,筛选出2份每角果粒数较多的油菜种质资源。基于GLM模型检测到39个与油菜每角果粒数显著关联SNPs,采用MLM分析发现,两地共检测到的3个每角果粒数显著关联SNPs位点均在GLM检测到。8个位点附近找到CIK,ERF022和EDE1等19个拟南芥已报道角果籽粒发育相关的同源基因。研究结果有助于解析甘蓝型油菜每角果粒数的遗传基础,为研究每角果粒数的调控机制、指导每角果粒数的遗传改良奠定基础。
肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151.
XIAO Xiao-jun, CHEN Ming, HAN De-peng, YU Pao-lan, ZHENG Wei, XIAO Guo-bin, ZHOU Qing-hong, ZHOU Hui-wen. Genome Wide Association Analysis of Thousand Seed Weight in Brassica napus L.[J]. Biotechnology Bulletin, 2023, 39(3): 143-151.
| 性状 Trait | 环境 Environment | 平均值±标准差 Mean±SD | 最小值 Min | 50%分位数 50% quantile | 最大值 Max | 变异系数 CV /% | Kolmogorov-Smirnov |
|---|---|---|---|---|---|---|---|
| 每角果粒数 SPS | JXAU | 17.23±2.89 | 7.40 | 17.41 | 24.48 | 16.77 | P=0.729 58 |
| JXIRS | 15.79±3.36 | 5.54 | 15.88 | 27.78 | 21.29 | P=0.346 01 |
表1 每角果粒数统计分析
Table 1 Statistical analysis on SPS(seed number per silique)
| 性状 Trait | 环境 Environment | 平均值±标准差 Mean±SD | 最小值 Min | 50%分位数 50% quantile | 最大值 Max | 变异系数 CV /% | Kolmogorov-Smirnov |
|---|---|---|---|---|---|---|---|
| 每角果粒数 SPS | JXAU | 17.23±2.89 | 7.40 | 17.41 | 24.48 | 16.77 | P=0.729 58 |
| JXIRS | 15.79±3.36 | 5.54 | 15.88 | 27.78 | 21.29 | P=0.346 01 |
| 变异来源Source of variation | 自由度DF(degree of freedom) | 方差及占总方差的比例MS(SS/SST)/% |
|---|---|---|
| 区组 Block | 1 | 52.12**(7.02) |
| 处理间 Treatment | 299 | 20.66**(2.78) |
| 环境 Environment/E | 1 | 621.99**(83.80) |
| 基因型 Genotype /G | 299 | 26.56**(3.58) |
| 基因型×环境 G×E | 299 | 12.75*(1.72) |
| 误差 Error | 599 | 8.17(1.10) |
表2 两地试验每角果粒数的方差分析
Table 2 Variance analysis of SPS in two places
| 变异来源Source of variation | 自由度DF(degree of freedom) | 方差及占总方差的比例MS(SS/SST)/% |
|---|---|---|
| 区组 Block | 1 | 52.12**(7.02) |
| 处理间 Treatment | 299 | 20.66**(2.78) |
| 环境 Environment/E | 1 | 621.99**(83.80) |
| 基因型 Genotype /G | 299 | 26.56**(3.58) |
| 基因型×环境 G×E | 299 | 12.75*(1.72) |
| 误差 Error | 599 | 8.17(1.10) |
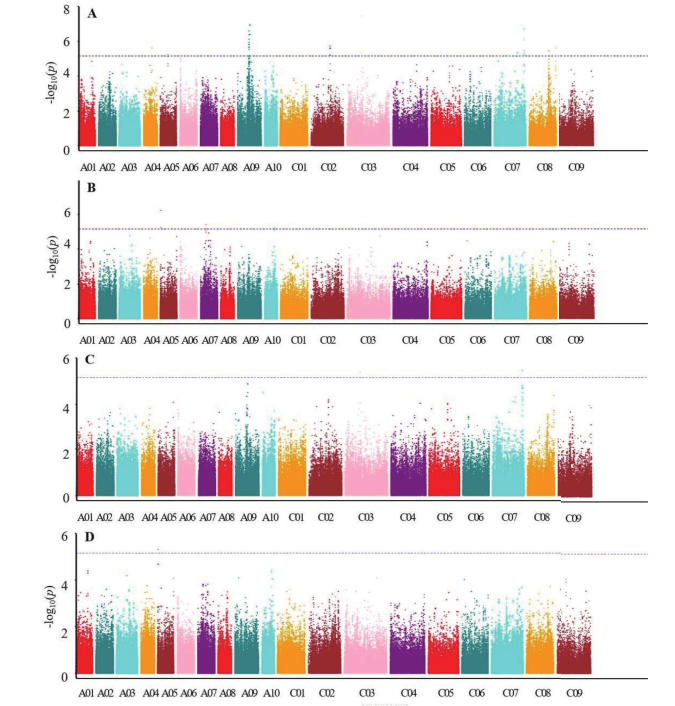
图2 每角果粒数全基因组关联分析Manhattant图 A:江西农业大学GLM;B:江西省红壤研究所GLM;C:江西农业大学MLM;D:江西省红壤研究所MLM
Fig. 2 Manhattan map of GWAS for SPS A:GLM in JXAU. B:GLM in JXIRS. C:MLM in JXAU. D:MLM in JXIRS
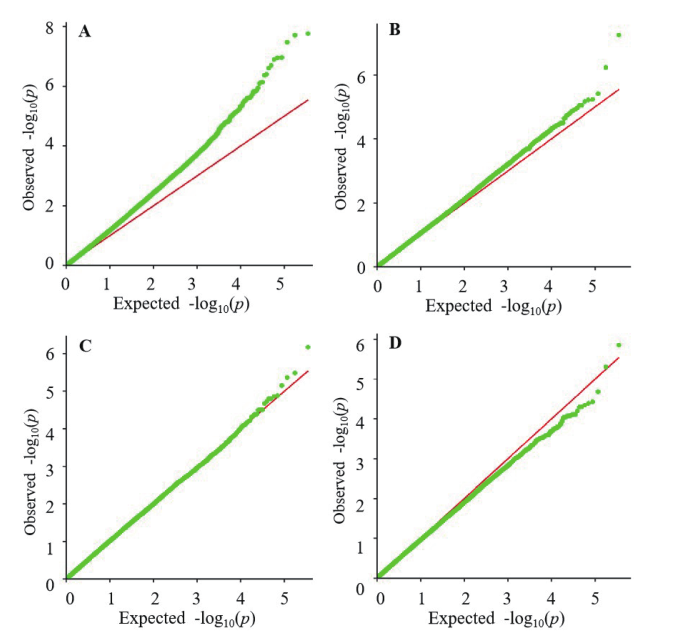
图3 每角果粒数全基因组关联分析Q-Q plot图 A:江西农业大学GLM;B:江西省红壤研究所GLM;C:江西农业大学MLM;D:江西省红壤研究所MLM
Fig. 3 Q-Q plot of GWAS for SPS A:GLM in JXAU.B:GLM in JXIRS.C:MLM in JXAU.D:MLM in JXIRS
| 环境Environment | 染色体 Chromosome | 位置 Locus | P值P value | 贡献率 R2/% | GLM | MLM |
|---|---|---|---|---|---|---|
| JXAU | A04 | 11830197 | 2.40E-06 | 11.07 | ● | |
| A05 | 10733464 | 2.32E-06 | 8.65 | ● | ||
| A09 | 16100373 | 1.11E-06 | 8.87 | ● | ||
| A09 | 16100374 | 1.55E-06 | 8.60 | ● | ||
| A09 | 16100386 | 1.65E-06 | 8.56 | ● | ||
| A09 | 16100422 | 1.39E-06 | 8.67 | ● | ||
| A09 | 16100439 | 1.35E-06 | 8.76 | ● | ||
| A09 | 16299217 | 1.68E-06 | 9.45 | ● | ||
| A09 | 16299389 | 1.49E-06 | 9.71 | ● | ||
| A09 | 16299462 | 1.13E-06 | 9.87 | ● | ||
| A09 | 16299495 | 4.31E-07 | 10.38 | ● | ||
| A09 | 16299626 | 4.29E-08 | 12.92 | ● | ||
| A09 | 16303740 | 3.33E-07 | 9.69 | ● | ||
| A09 | 16303768 | 1.98E-06 | 8.59 | ● | ||
| A09 | 16366473 | 2.84E-06 | 9.82 | ● | ||
| A09 | 17453710 | 1.10E-07 | 17.80 | ● | ||
| A09 | 17453729 | 1.29E-07 | 17.42 | ● | ||
| A09 | 17453891 | 4.02E-07 | 16.69 | ● | ||
| A09 | 17471305 | 2.81E-06 | 8.66 | ● | ||
| A09 | 17581807 | 3.44E-06 | 9.45 | ● | ||
| A09 | 17581835 | 3.73E-06 | 9.77 | ● | ||
| A09 | 17582057 | 3.16E-06 | 8.96 | ● | ||
| C02 | 27389727 | 2.58E-06 | 7.91 | ● | ||
| C02 | 27389750 | 1.90E-06 | 8.06 | ● | ||
| C02 | 27389775 | 2.39E-06 | 7.93 | ● | ||
| C02 | 34901810 | 4.35E-06 | 8.04 | ● | ||
| C03 | 18955107 | 4.53E-06 | 8.52 | ● | ||
| C03 | 21284972 | 3.40E-08-4.29E-06 | 10.85-8.9 | ● | ○ | |
| C03 | 21342259 | 4.40E-06 | 7.67 | ● | ||
| C07 | 32557367 | 4.69E-06 | 7.48 | ● | ||
| C07 | 42323087 | 8.09E-07 | 14.77 | ● | ||
| C07 | 42323117 | 2.00E-07-3.28E-06 | 15.23-14.05 | ● | ○ | |
| C07 | 42323137 | 3.24E-06 | 12.97 | ● | ||
| C07 | 42323279 | 4.80E-06 | 11.83 | ● | ||
| C07 | 42323280 | 4.90E-06 | 11.88 | ● | ||
| C08 | 27721260 | 3.47E-06 | 8.13 | ● | ||
| C08 | 37461613 | 2.43E-06 | 11.73 | ● | ||
| JXIRS | A05 | 659981 | 5.92E-07-4.95E-06 | 16.00-16.75 | ● | ○ |
| A07 | 7482038 | 3.89E-06 | 9.63 | ● |
表3 每角果粒数显著关联SNPs位点
Table 3 SNPs loci significantly associated with SPS
| 环境Environment | 染色体 Chromosome | 位置 Locus | P值P value | 贡献率 R2/% | GLM | MLM |
|---|---|---|---|---|---|---|
| JXAU | A04 | 11830197 | 2.40E-06 | 11.07 | ● | |
| A05 | 10733464 | 2.32E-06 | 8.65 | ● | ||
| A09 | 16100373 | 1.11E-06 | 8.87 | ● | ||
| A09 | 16100374 | 1.55E-06 | 8.60 | ● | ||
| A09 | 16100386 | 1.65E-06 | 8.56 | ● | ||
| A09 | 16100422 | 1.39E-06 | 8.67 | ● | ||
| A09 | 16100439 | 1.35E-06 | 8.76 | ● | ||
| A09 | 16299217 | 1.68E-06 | 9.45 | ● | ||
| A09 | 16299389 | 1.49E-06 | 9.71 | ● | ||
| A09 | 16299462 | 1.13E-06 | 9.87 | ● | ||
| A09 | 16299495 | 4.31E-07 | 10.38 | ● | ||
| A09 | 16299626 | 4.29E-08 | 12.92 | ● | ||
| A09 | 16303740 | 3.33E-07 | 9.69 | ● | ||
| A09 | 16303768 | 1.98E-06 | 8.59 | ● | ||
| A09 | 16366473 | 2.84E-06 | 9.82 | ● | ||
| A09 | 17453710 | 1.10E-07 | 17.80 | ● | ||
| A09 | 17453729 | 1.29E-07 | 17.42 | ● | ||
| A09 | 17453891 | 4.02E-07 | 16.69 | ● | ||
| A09 | 17471305 | 2.81E-06 | 8.66 | ● | ||
| A09 | 17581807 | 3.44E-06 | 9.45 | ● | ||
| A09 | 17581835 | 3.73E-06 | 9.77 | ● | ||
| A09 | 17582057 | 3.16E-06 | 8.96 | ● | ||
| C02 | 27389727 | 2.58E-06 | 7.91 | ● | ||
| C02 | 27389750 | 1.90E-06 | 8.06 | ● | ||
| C02 | 27389775 | 2.39E-06 | 7.93 | ● | ||
| C02 | 34901810 | 4.35E-06 | 8.04 | ● | ||
| C03 | 18955107 | 4.53E-06 | 8.52 | ● | ||
| C03 | 21284972 | 3.40E-08-4.29E-06 | 10.85-8.9 | ● | ○ | |
| C03 | 21342259 | 4.40E-06 | 7.67 | ● | ||
| C07 | 32557367 | 4.69E-06 | 7.48 | ● | ||
| C07 | 42323087 | 8.09E-07 | 14.77 | ● | ||
| C07 | 42323117 | 2.00E-07-3.28E-06 | 15.23-14.05 | ● | ○ | |
| C07 | 42323137 | 3.24E-06 | 12.97 | ● | ||
| C07 | 42323279 | 4.80E-06 | 11.83 | ● | ||
| C07 | 42323280 | 4.90E-06 | 11.88 | ● | ||
| C08 | 27721260 | 3.47E-06 | 8.13 | ● | ||
| C08 | 37461613 | 2.43E-06 | 11.73 | ● | ||
| JXIRS | A05 | 659981 | 5.92E-07-4.95E-06 | 16.00-16.75 | ● | ○ |
| A07 | 7482038 | 3.89E-06 | 9.63 | ● |
| 基因ID Gene ID | SNP 标记 SNP marker | 起点 Gene start | 终点 Gene end | SNP 位置 SNP position | 拟南芥基因号Arabidopsis gene ID | 拟南芥同源基因 Arabidopsis alias |
|---|---|---|---|---|---|---|
| BnaA04g14020D | Bn-A04-11830197 | 11833699 | 11837146 | 11830197 | AT2G23950 | CIK[ |
| BnaA09g23540D | Bn-A09-16100373 | 16174364 | 16174915 | 16100373 | AT1G33760 | ERF022[ |
| BnaA09g24700D | Bn-A09-17471305 | 17487033 | 17488935 | 17471305 | AT2G44190 | EDE1[ |
| BnaA09g24710D | Bn-A09-17471305 | 17497234 | 17500297 | 17471305 | AT1G31930 | XLG3[ |
| BnaA09g24730D | Bn-A09-17471305 | 17523243 | 17523516 | 17471305 | AT1G31870 | AtBUD13[ |
| BnaA09g24750D | Bn-A09-17471305 | 17541993 | 17543181 | 17471305 | AT4G34200 | EDA9[ |
| BnaA09g24870D | Bn-A09-17581807 | 17647230 | 17648587 | 17581807 | AT1G31817 | NFD3[ |
| BnaC03g34930D | Bn-C03-21284972 | 21219565 | 21222207 | 21284972 | AT3G06830 | PME[ |
| BnaC03g34950D | Bn-C03-21284972 | 21230822 | 21235581 | 21284972 | AT3G06860 | MFP2[ |
| BnaC03g35050D | Bn-C03-21284972 | 21284496 | 21287359 | 21284972 | AT3G07050 | NSN1[ |
| BnaC03g35090D | Bn-C03-21284972 | 21319583 | 21320097 | 21284972 | AT5G52820 | NLE[ |
| BnaC03g35120D | Bn-C03-21284972 | 21340067 | 21357495 | 21284972 | AT3G07160 | GSL10[ |
| BnaC03g35200D | Bn-C03-21284972 | 21380681 | 21384039 | 21284972 | AT3G07330 | CSLC6[ |
| BnaC07g26700D | Bn-C07-32557367 | 32520784 | 32523680 | 32557367 | AT5G48720 | XRI1[ |
| BnaC07g26770D | Bn-C07-32557367 | 32544716 | 32546391 | 32557367 | AT5G48820 | ICK6/KRP3[ |
| BnaC07g27010D | Bn-C07-32557367 | 32640586 | 32647933 | 32557367 | AT5G49030 | OVA2[ |
| BnaC08g26210D | Bn-C08-27721260 | 27683429 | 27686693 | 27721260 | AT3G55400 | OVA1[ |
| BnaC08g26310D | Bn-C08-27721260 | 27766812 | 27771782 | 27721260 | AT3G55480 | AP-3[ |
| BnaC08g44100D | Bn-C08-37461613 | 37532295 | 37541481 | 37461613 | AT1G05230 | HDG2[ |
表4 每角果粒数显著关联SNP标记区间候选基因
Table 4 Candidate genes related with SPS in the intervals of significant associated SNP markers
| 基因ID Gene ID | SNP 标记 SNP marker | 起点 Gene start | 终点 Gene end | SNP 位置 SNP position | 拟南芥基因号Arabidopsis gene ID | 拟南芥同源基因 Arabidopsis alias |
|---|---|---|---|---|---|---|
| BnaA04g14020D | Bn-A04-11830197 | 11833699 | 11837146 | 11830197 | AT2G23950 | CIK[ |
| BnaA09g23540D | Bn-A09-16100373 | 16174364 | 16174915 | 16100373 | AT1G33760 | ERF022[ |
| BnaA09g24700D | Bn-A09-17471305 | 17487033 | 17488935 | 17471305 | AT2G44190 | EDE1[ |
| BnaA09g24710D | Bn-A09-17471305 | 17497234 | 17500297 | 17471305 | AT1G31930 | XLG3[ |
| BnaA09g24730D | Bn-A09-17471305 | 17523243 | 17523516 | 17471305 | AT1G31870 | AtBUD13[ |
| BnaA09g24750D | Bn-A09-17471305 | 17541993 | 17543181 | 17471305 | AT4G34200 | EDA9[ |
| BnaA09g24870D | Bn-A09-17581807 | 17647230 | 17648587 | 17581807 | AT1G31817 | NFD3[ |
| BnaC03g34930D | Bn-C03-21284972 | 21219565 | 21222207 | 21284972 | AT3G06830 | PME[ |
| BnaC03g34950D | Bn-C03-21284972 | 21230822 | 21235581 | 21284972 | AT3G06860 | MFP2[ |
| BnaC03g35050D | Bn-C03-21284972 | 21284496 | 21287359 | 21284972 | AT3G07050 | NSN1[ |
| BnaC03g35090D | Bn-C03-21284972 | 21319583 | 21320097 | 21284972 | AT5G52820 | NLE[ |
| BnaC03g35120D | Bn-C03-21284972 | 21340067 | 21357495 | 21284972 | AT3G07160 | GSL10[ |
| BnaC03g35200D | Bn-C03-21284972 | 21380681 | 21384039 | 21284972 | AT3G07330 | CSLC6[ |
| BnaC07g26700D | Bn-C07-32557367 | 32520784 | 32523680 | 32557367 | AT5G48720 | XRI1[ |
| BnaC07g26770D | Bn-C07-32557367 | 32544716 | 32546391 | 32557367 | AT5G48820 | ICK6/KRP3[ |
| BnaC07g27010D | Bn-C07-32557367 | 32640586 | 32647933 | 32557367 | AT5G49030 | OVA2[ |
| BnaC08g26210D | Bn-C08-27721260 | 27683429 | 27686693 | 27721260 | AT3G55400 | OVA1[ |
| BnaC08g26310D | Bn-C08-27721260 | 27766812 | 27771782 | 27721260 | AT3G55480 | AP-3[ |
| BnaC08g44100D | Bn-C08-37461613 | 37532295 | 37541481 | 37461613 | AT1G05230 | HDG2[ |
| [1] | 王汉中. 我国油菜产业发展的历史回顾与展望[J]. 中国油料作物学报, 2010, 32(2): 300-302. |
| Wang HZ. Review and future development of rapeseed industry in China[J]. Chin J Oil Crop Sci, 2010, 32(2): 300-302. | |
| [2] | 王汉中. 以新需求为导向的油菜产业发展战略[J]. 中国油料作物学报, 2018, 40(5): 613-617. |
| Wang HZ. New-demand oriented oilseed rape industry developing strategy[J]. Chin J Oil Crop Sci, 2018, 40(5): 613-617. | |
| [3] | 刘成, 冯中朝, 肖唐华, 等. 我国油菜产业发展现状、潜力及对策[J]. 中国油料作物学报, 2019, 41(4): 485-489. |
| Liu C, Feng ZC, Xiao TH, et al. Development, potential and adaptation of Chinese rapeseed industry[J]. Chin J Oil Crop Sci, 2019, 41(4): 485-489. | |
| [4] |
Van CW. Yield enhancement genes: seeds for growth[J]. Curr Opin Biotechnol, 2005, 16(2): 147-153.
doi: 10.1016/j.copbio.2005.03.002 URL |
| [5] |
Chen W, Zhang Y, Liu XP, et al. Detection of QTL for six yield-related traits in oilseed rape(Brassica napus)using DH and immortalized F2 populations[J]. Theor Appl Genet, 2007, 115(6): 849-858.
doi: 10.1007/s00122-007-0613-2 pmid: 17665168 |
| [6] |
Fan CC, Cai GQ, Qin J, et al. Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus[J]. Theor Appl Genet, 2010, 121(7): 1289-1301.
doi: 10.1007/s00122-010-1388-4 URL |
| [7] | Zhu LX, Zhang DX, Fu TD, et al. Analysis of yield and disease resistance traits of new winter rapeseed varieties over the past twenty years in China[J]. Agric Sci & Technol, 2011, 12(6): 842-846. |
| [8] | 易斌, 陈伟, 马朝芝, 等. 甘蓝型油菜产量及相关性状的QTL分析[J]. 作物学报, 2006, 32(5): 676-682. |
| Yi B, Chen W, Ma CZ, et al. Mapping of quantitative trait loci for yield and yield components in Brassica napus L[J]. Acta Agron Sin, 2006, 32(5): 676-682. | |
| [9] | 张月立. 甘蓝型油菜含油量、角果长和每果粒数的QTL定位[D]. 武汉: 华中农业大学, 2013. |
| Zhang YL. Qtl mapping for oil content, silique length and seed number per silique in Brassica napus L[D]. Wuhan: Huazhong Agricultural University, 2013. | |
| [10] | 赵卫国, 王灏, 穆建新, 等. 甘蓝型油菜千粒重性状的QTL定位分析[J]. 西北植物学报, 2017, 37(3): 478-485. |
| Zhao WG, Wang H, Mu JX, et al. Localization of thousand seed weight trait in Brassica napus by quantitative trait locus analysis[J]. Acta Bot Boreali Occidentalia Sin, 2017, 37(3): 478-485. | |
| [11] |
孙程明, 陈松, 彭琦, 等. 甘蓝型油菜角果长度性状的全基因组关联分析[J]. 作物学报, 2019, 45(9): 1303-1310.
doi: 10.3724/SP.J.1006.2019.94021 |
| Sun CM, Chen S, Peng Q, et al. Genome-wide association study of silique length in rapeseed(Brassica napus L.)[J]. Acta Agron Sin, 2019, 45(9): 1303-1310. | |
| [12] | 王茹梦, 刘忠松. 甘蓝型油菜开花期全基因组关联分析及开花基因标记开发[J]. 分子植物育种, 2021, 19(10): 3329-3338. |
| Wang RM, Liu ZS. Genome-wide association analysis of flowering time and development of flowering gene markers in Brassica napus L[J]. Mol Plant Breed, 2021, 19(10): 3329-3338. | |
| [13] |
周庆红, 周灿, 郑伟, 等. 甘蓝型油菜角果长度全基因组关联分析[J]. 中国农业科学, 2017, 50(2): 228-239.
doi: 10.3864/j.issn.0578-1752.2017.02.003 |
| Zhou QH, Zhou C, Zheng W, et al. Genome wide association analysis of silique length in Brassica napus L[J]. Sci Agric Sin, 2017, 50(2): 228-239. | |
| [14] |
韩德鹏, 周灿, 郑伟, 等. 甘蓝型油菜主花序性状全基因组关联分析[J]. 核农学报, 2018, 32(3): 463-476.
doi: 10.11869/j.issn.100-8551.2018.03.0463 |
| Han DP, Zhou C, Zheng W, et al. Genome wide association analysis of main inflorescence related traits in Brassica napus L[J]. J Nucl Agric Sci, 2018, 32(3): 463-476. | |
| [15] |
Zhao XZ, Yu KJ, Pang CK, et al. QTL analysis of five silique-related traits in Brassica napus L. across multiple environments[J]. Front Plant Sci, 2021, 12: 766271.
doi: 10.3389/fpls.2021.766271 URL |
| [16] |
Wang XD, Chen L, Wang AN, et al. Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus[J]. BMC Plant Biol, 2016, 16: 71.
doi: 10.1186/s12870-016-0759-7 URL |
| [17] | 任义英, 崔翠, 王倩, 等. 甘蓝型油菜籽粒着生密度及其相关性状全基因组关联分析[J]. 植物遗传资源学报, 2018, 19(2): 314-325. |
| Ren YY, Cui C, Wang Q, et al. Genome-wide association analysis of seed density within per silique and its related traits in Brassica napus L[J]. J Plant Genet Resour, 2018, 19(2): 314-325. | |
| [18] |
Zhou QH, Zhou C, Zheng W, et al. Genome-wide SNP markers based on SLAF-seq uncover breeding traces in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2017, 8: 648.
doi: 10.3389/fpls.2017.00648 URL |
| [19] |
Sun XW, Liu DY, Zhang XF, et al. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing[J]. PLoS One, 2013, 8(3): e58700.
doi: 10.1371/journal.pone.0058700 URL |
| [20] | 伍晓明, 陈碧云, 陆光远. 油菜种质资源描述规范和数据标准[M]. 北京: 中国农业出版社, 2007. |
| Wu XM, Chen BY, Lu GY. Descriptors and data standard for rapeseed(Brassica spp.)[M]. Beijing: Chinese Agriculture Press, 2007. | |
| [21] |
Cui YW, Hu C, Zhu YF, et al. CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis[J]. Plant Cell, 2018, 30(10): 2383-2401.
doi: 10.1105/tpc.17.00586 URL |
| [22] |
Nowak K, Wójcikowska B, Gaj MD. ERF022 impacts the induction of somatic embryogenesis in Arabidopsis through the ethylene-related pathway[J]. Planta, 2015, 241(4): 967-985.
doi: 10.1007/s00425-014-2225-9 URL |
| [23] |
Pignocchi C, Minns GE, Nesi N, et al. ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis[J]. Plant Cell, 2009, 21(1): 90-105.
doi: 10.1105/tpc.108.061812 pmid: 19151224 |
| [24] |
Wang YP, Wu YY, Yu BY, et al. EXTRA-LARGE G PROTEINs interact with E3 ligases PUB4 and PUB2 and function in cytokinin and developmental processes[J]. Plant Physiol, 2017, 173(2): 1235-1246.
doi: 10.1104/pp.16.00816 pmid: 27986866 |
| [25] |
Xiong F, Ren JJ, Yu Q, et al. AtBUD13 affects pre-mRNA splicing and is essential for embryo development in Arabidopsis[J]. Plant J, 2019, 98(4): 714-726.
doi: 10.1111/tpj.14268 |
| [26] |
Toujani W, Muñoz-Bertomeu J, Flores-Tornero M, et al. Identification of the phosphoglycerate dehydrogenase isoform EDA9 as the essential gene for embryo and male gametophyte development in Arabidopsis[J]. Plant Signal Behav, 2013, 8(11): e27207.
doi: 10.4161/psb.27207 URL |
| [27] |
Portereiko MF, Sandaklie-Nikolova L, Lloyd A, et al. NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization[J]. Plant Physiol, 2006, 141(3): 957-965.
doi: 10.1104/pp.106.079319 pmid: 16698901 |
| [28] |
Louvet R, Cavel E, Gutierrez L, et al. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana[J]. Planta, 2006, 224(4): 782-791.
doi: 10.1007/s00425-006-0261-9 URL |
| [29] |
Richmond TA, Bleecker AB. A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis[J]. Plant Cell, 1999, 11(10): 1911-1924.
pmid: 10521521 |
| [30] |
Wang XM, Xie B, Zhu MS, et al. Nucleostemin-like 1 is required for embryogenesis and leaf development in Arabidopsis[J]. Plant Mol Biol, 2012, 78(1-2): 31-44.
doi: 10.1007/s11103-011-9840-7 URL |
| [31] |
Chantha SC, Gray-Mitsumune M, Houde J, et al. The MIDASIN and NOTCHLESS genes are essential for female gametophyte development in Arabidopsis thaliana[J]. Physiol Mol Biol Plants, 2010, 16(1): 3-18.
doi: 10.1007/s12298-010-0005-y URL |
| [32] |
Töller A, Brownfield L, Neu C, et al. Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth[J]. Plant J, 2008, 54(5): 911-923.
doi: 10.1111/j.1365-313X.2008.03462.x URL |
| [33] |
Boavida LC, Shuai B, Yu HJ, et al. A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana[J]. Genetics, 2009, 181(4): 1369-1385.
doi: 10.1534/genetics.108.090852 pmid: 19237690 |
| [34] |
Dean PJ, Siwiec T, Waterworth WM, et al. A novel ATM-dependent X-ray-inducible gene is essential for both plant meiosis and gametogenesis[J]. Plant J, 2009, 58(5): 791-802.
doi: 10.1111/tpj.2009.58.issue-5 URL |
| [35] |
Cheng Y, Cao L, Wang S, et al. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis[J]. Plant J, 2013, 75(4): 642-655.
doi: 10.1111/tpj.2013.75.issue-4 URL |
| [36] |
Berg M, Rogers R, Muralla R, et al. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis[J]. Plant J, 2005, 44(5): 866-878.
doi: 10.1111/tpj.2005.44.issue-5 URL |
| [37] |
Feng QN, Liang X, Li S, et al. The ADAPTOR PROTEIN-3 complex mediates pollen tube growth by coordinating vacuolar targeting and organization[J]. Plant Physiol, 2018, 177(1): 216-225.
doi: 10.1104/pp.17.01722 URL |
| [38] |
Kamata N, Okada H, Komeda Y, et al. Mutations in epidermis-specific HD-ZIP IV genes affect floral organ identity in Arabidopsis thaliana[J]. Plant J, 2013, 75(3): 430-440.
doi: 10.1111/tpj.2013.75.issue-3 URL |
| [39] | Ahmad A. 甘蓝型油菜初始胚珠数和每角果粒数数量性状位点(QTL)的研究[D]. 武汉: 华中农业大学, 2018. |
| Ahmad A. Quantitative trait loci(QTL)studies for the number of ovules and seeds per pod in Brassica napus L.[D]. Wuhan: Huazhong Agricultural University, 2018. | |
| [40] |
钟婷婷, 郭诗芬, 卢文斌, 等. 甘蓝型油菜抗根肿病KASP标记开发和利用[J]. 华北农学报, 2021, 36(4): 184-190.
doi: 10.7668/hbnxb.20192042 |
| Zhong TT, Guo SF, Lu WB, et al. Development and utilization of KASP marker for Brassica napus clubroot resistance breeding[J]. Acta Agric Boreali Sin, 2021, 36(4): 184-190. | |
| [41] |
Jiao YM, Zhang KP, Cai GQ, et al. Fine mapping and candidate gene analysis of a major locus controlling ovule abortion and seed number per silique in Brassica napus L[J]. Theor Appl Genet, 2021, 134(8): 2517-2530.
doi: 10.1007/s00122-021-03839-6 |
| [42] |
Zhai YG, Cai SL, Hu LM, et al. CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L[J]. Theor Appl Genet, 2019, 132(7): 2111-2123.
doi: 10.1007/s00122-019-03341-0 |
| [43] |
Zheng M, Zhang L, Tang M, et al. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed(Brassica napus L.)[J]. Plant Biotechnol J, 2020, 18(3): 644-654.
doi: 10.1111/pbi.13228 pmid: 31373135 |
| [44] | 高谢旺, 谭安琪, 胡信畅, 等. 利用CRISPR/Cas9技术创制高油酸甘蓝型油菜新种质[J]. 植物遗传资源学报, 2020, 21(4): 1002-1008. |
| Gao XW, Tan AQ, Hu XC, et al. Creation of new germplasm of high-oleic rapeseed using CRISPR/Cas9[J]. J Plant Genet Resour, 2020, 21(4): 1002-1008. |
| [1] | 蔡梦鲜, 高作敏, 胡利娟, 冯群, 王洪程, 朱斌. 天然甘蓝型油菜C染色体组C1,C2缺体的创建及遗传分析[J]. 生物技术通报, 2023, 39(3): 81-88. |
| [2] | 支添添, 周舟, 陈纪鹏, 韩成云. 甘蓝型油菜酪氨酸代谢关键基因FAH的克隆、功能鉴定和表达分析[J]. 生物技术通报, 2023, 39(10): 115-127. |
| [3] | 金姣姣, 刘自刚, 米文博, 徐明霞, 邹娅, 徐春梅, 赵彩霞. 利用RNA-Seq鉴定调控甘蓝型油菜叶片光合特性的低温胁迫应答基因[J]. 生物技术通报, 2022, 38(4): 126-142. |
| [4] | 刘世男, 林新春. 植物SVP基因的研究进展[J]. 生物技术通报, 2014, 0(6): 9-13. |
| [5] | 郝晓云, 沈海涛, 李鸿彬. 甘蓝型油菜下胚轴和带柄子叶再生体系研究[J]. 生物技术通报, 2013, 0(4): 69-74. |
| [6] | 杨长友, 袁中厚, 郑小敏, 张涛. 甘蓝型油菜高效离体再生体系的建立[J]. 生物技术通报, 2013, 0(1): 111-115. |
| [7] | 孔芳;薛正莲;杨超英;王幼平;. 转Bar基因甘蓝型油菜叶片蛋白质组变化的初步分析[J]. , 2012, 0(10): 75-82. |
| [8] | 姜嵛;梁晨;姜国勇;. 植物内生枯草芽孢杆菌纳豆激酶同源基因的序列分析及其在大肠杆菌中的表达[J]. , 2011, 0(03): 97-99. |
| [9] | 邓秋红;栗茂腾;向福;余龙江;. 甘蓝型油菜β碳酸酐酶的结构预测[J]. , 2010, 0(01): 111-117. |
| [10] | 刘建民;李运涛;甘露;李红;栗茂腾;. napin基因启动子克隆及进化分析[J]. , 2008, 0(S1): 188-191. |
| [11] | 邹智;吴刚;肖玲;武玉花;聂淑晶;肖娜;卢长明;. 植物源内含子对GUS基因表达模式的影响[J]. , 2008, 0(06): 78-82. |
| [12] | 秦春圃;. 甘蓝型油菜BnKCR2基因cDNA的克隆和真核表达载体的构建[J]. , 2008, 0(04): 201-201. |
| [13] | 路小春;李加纳;李本逊;柴友荣;. 甘蓝型油菜TT1基因反义植物表达载体的构建[J]. , 2006, 0(S1): 305-309. |
| [14] | 秦春圃. 应用RAPD标记法鉴定甘蓝型油菜杂种纯度[J]. , 2003, 0(04): 54-54. |
| [15] | 孙雷心. 发现“绿色革命”基因[J]. , 1999, 0(06): 52-52. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||