








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 133-142.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0690
收稿日期:2022-06-04
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
宋丽,女,博士,教授,研究方向:作物遗传育种;E-mail: songli@yzu.edu.cn作者简介:杨春洪,女,硕士研究生,研究方向:大豆功能基因组学;E-mail: azneych@outlook.com
基金资助:
YANG Chun-hong( ), DONG Lu, CHEN Lin, SONG Li(
), DONG Lu, CHEN Lin, SONG Li( )
)
Received:2022-06-04
Published:2023-03-26
Online:2023-04-10
摘要:
拟南芥VAS1基因编码一个磷酸吡哆醛依赖性氨基转移酶,可将吲哚丙酮酸转化为吲哚乙酸的生物合成前体色氨酸,是生长素代谢调控中的一个关键酶。对大豆VAS1基因家族进行全基因组鉴定与表达模式分析,并探究其在根系发育中的作用,为深入挖掘大豆VAS1基因的功能奠定基础。运用生物信息学方法对该基因家族成员进行鉴定和分析,采用实时荧光定量分析该家族在不同逆境处理下的表达模式,克隆GmVAS1-1,进一步研究其功能,并通过酵母双杂交试验筛选大豆GmVAS1-1蛋白的互作因子。结果表明,大豆VAS1基因家族共有2个成员,命名为GmVAS1-1和GmVAS1-2。系统进化树、基因结构和保守基序分析表明,大豆VAS1与豆科植物亲缘关系更近。启动子序列分析发现多个逆境和光响应元件。表达模式分析表明,大豆VAS1家族基因在不同种子发育时期的胚乳中表达量较高。荧光定量PCR分析表明,在干旱和盐胁迫下,大豆根系VAS1基因表达量显著上调。拟南芥过量表达GmVAS1-1植株侧根数较野生型显著减少。酵母互作试验及预测共筛选到17个与GmVAS1-1互作的蛋白。大豆VAS1基因家族成员在胚乳中表达量较高,且响应多种逆境及营养胁迫,其中,GmVAS1-1参与侧根发育。
杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142.
YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development[J]. Biotechnology Bulletin, 2023, 39(3): 133-142.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| GmVAS1-AscI-F | GGCGCGCCCTCCCGCAGTCCGTCTCT |
| GmVAS1-BamHI-R | GGATCCATGGGTTCGTTCGTGAAG |
| AT1G80360-LP | TCTTGAATCCAATTCCGTGAC |
| AT1G80360-RP | CTTAACGTTCTCCTTCCCCAG |
| LB2 | GCTTCCTATTATATCTTCCCAAATTACCAATACA |
| AtActin-qPCR-F | TACCGAGGCTCCTCTTAACC |
| AtActin-qPCR-R | AGCTTGGATGGCGACATACA |
| AT1G80360-qPCR-F | CCAAGTGGCACCTATGTCC |
| AT1G80360-qPCR-R | ACAATGTGGTCTCCCTCAAC |
| GmVAS1-NdeI-F | CCCATATGATGGGTTCGTTCGTGAAG |
| GmVAS1-PstI-R | AACTGCAGTTACTCCCGCAGTCCGTCT |
| GmActin11-qPCR-F | ATCTTGACTGAGCGTGGTTATTCC |
| GmActin11- qPCR-R | GCTGGTCCTGGCTGTCTCC |
| GmVAS1-1- qPCR-F | TGATGGCCTGAAACACTCTTG |
| GmVAS1-1- qPCR-R | GCAAAGTCTTTTACTTCAGAGGG |
| GmVAS1-2- qPCR-F | CAAAGCTCCCAGACTTAGACG |
| GmVAS1-2- qPCR-R | TGCAGTCATTCTCCGTCAAG |
| pGBKT7-T7 | TAATACGACTCACTATAGGGCGAGC |
| pGBKT7-ADR | GTGAACTTGCGGGGTTTTTCAGTAT |
表1 引物序列
Table 1 Primer sequence
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| GmVAS1-AscI-F | GGCGCGCCCTCCCGCAGTCCGTCTCT |
| GmVAS1-BamHI-R | GGATCCATGGGTTCGTTCGTGAAG |
| AT1G80360-LP | TCTTGAATCCAATTCCGTGAC |
| AT1G80360-RP | CTTAACGTTCTCCTTCCCCAG |
| LB2 | GCTTCCTATTATATCTTCCCAAATTACCAATACA |
| AtActin-qPCR-F | TACCGAGGCTCCTCTTAACC |
| AtActin-qPCR-R | AGCTTGGATGGCGACATACA |
| AT1G80360-qPCR-F | CCAAGTGGCACCTATGTCC |
| AT1G80360-qPCR-R | ACAATGTGGTCTCCCTCAAC |
| GmVAS1-NdeI-F | CCCATATGATGGGTTCGTTCGTGAAG |
| GmVAS1-PstI-R | AACTGCAGTTACTCCCGCAGTCCGTCT |
| GmActin11-qPCR-F | ATCTTGACTGAGCGTGGTTATTCC |
| GmActin11- qPCR-R | GCTGGTCCTGGCTGTCTCC |
| GmVAS1-1- qPCR-F | TGATGGCCTGAAACACTCTTG |
| GmVAS1-1- qPCR-R | GCAAAGTCTTTTACTTCAGAGGG |
| GmVAS1-2- qPCR-F | CAAAGCTCCCAGACTTAGACG |
| GmVAS1-2- qPCR-R | TGCAGTCATTCTCCGTCAAG |
| pGBKT7-T7 | TAATACGACTCACTATAGGGCGAGC |
| pGBKT7-ADR | GTGAACTTGCGGGGTTTTTCAGTAT |
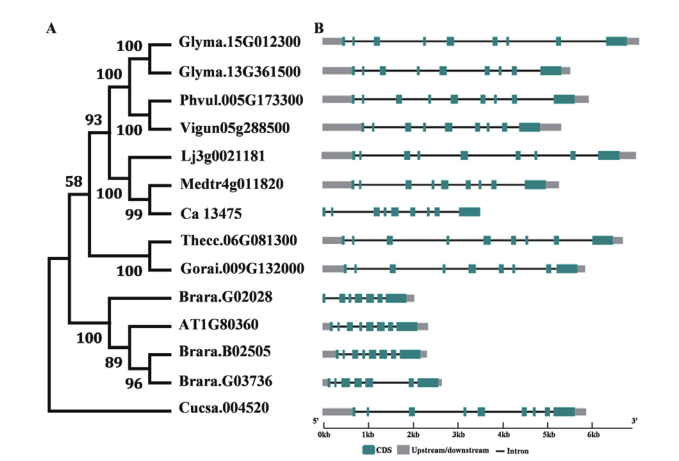
图1 VAS1基因家族进化分析和基因结构 大豆(Glycine max):Glyma.15G012300, Glyma.13G361500;菜豆(Phaseolus vulgaris):Phvul.005G173300;豇豆(Vigna unguiculata):Vigun05g288500;百脉根(Lotus corniculatus):Lj3g0021181;苜蓿(Medicago truncatula):Med-tr4g011820;鹰嘴豆(Cicer arietinum):Ca13475;烟草(Nicotiana tabacum):The.06G081300;雷蒙德氏棉(Gossypium raimondii):Gorai.009G132000;油菜(Brassica campestris):Brara.G02028, Brara.B02505, Brara.G03736;拟南芥(Arabidopsis thaliana):AT1G80360;栽培黄瓜(Cucumis sativus):Cucsa.004520
Fig. 1 Phylogenetic tree and gene structure of VAS1 gene family
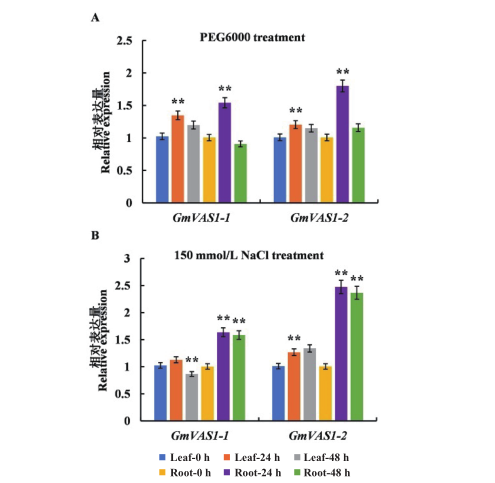
图5 PEG和NaCl处理下GmVAS1基因家族的表达模式 *:P<0.05;**:P<0.01。下同
Fig. 5 Expression pattern of GmVAS1 gene family under PEG or NaCl treatment *: P<0.05; **: P<0.01. The same below
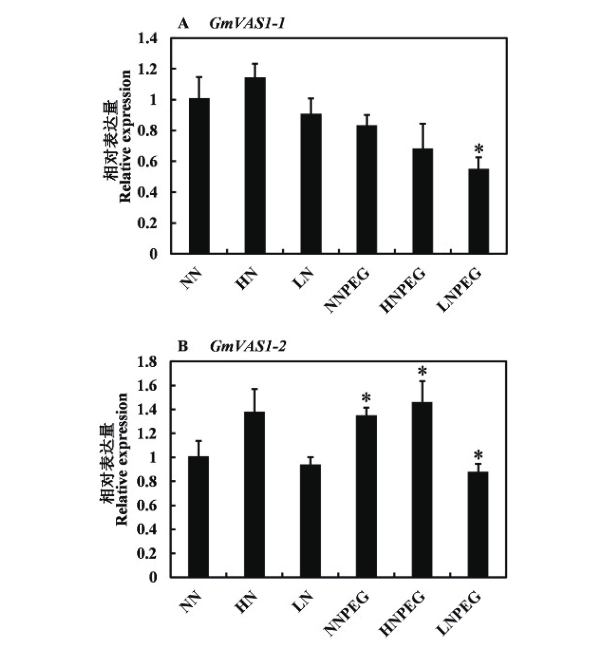
图6 不同浓度硝酸盐和PEG协同处理下GmVAS1表达模式 NN:适宜浓度硝酸盐;HN:高浓度硝酸盐;LN:低浓度硝酸盐;NNPEG:适宜浓度硝酸盐和PEG复合处理;HNPEG:高浓度硝酸盐和PEG复合处理;LNPEG:低浓度硝酸盐和PEG复合处理
Fig. 6 GmVAS1 expression patterns under co-treatment of different concentrations of nitrate and PEG NN: Normal concentration nitrate. HN: High concentration nitrate. LN: Low concentration nitrate. NNPEG: Normal concentration nitrate and PEG composite treatment. HNPEG: High concentration nitrate and PEG composite treatment. LNPEG: Low concentration nitrate and PEG composite treatment
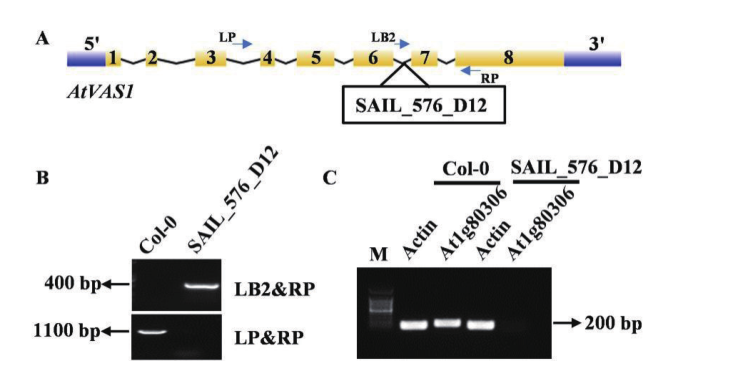
图7 拟南芥vas1突变体鉴定 A:T-DNA插入位置;B:T-DNA插入鉴定;C:RNA水平分析
Fig. 7 Mutant identification vas1 in Arabidopsis thaliana A: T-DNA insertion location in vas1. B: T-DNA insertion identification. C: RNA level identification
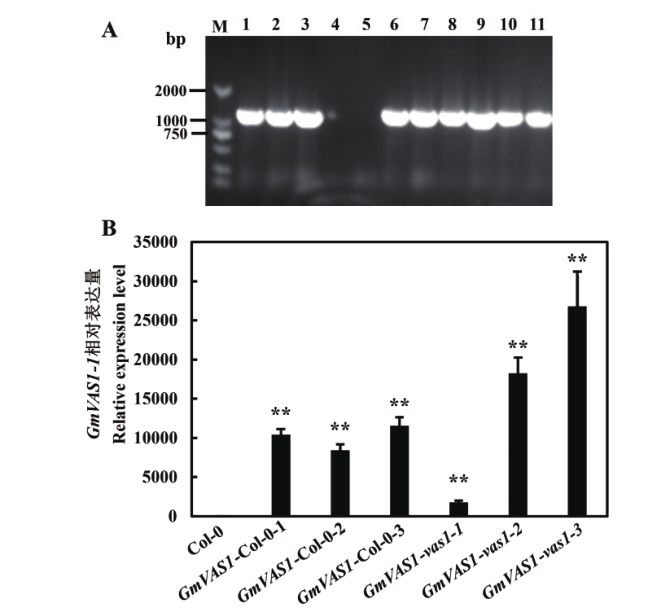
图8 拟南芥转基因植株鉴定 A:转基因拟南芥阳性植株筛选(M:DNA分子标准;1:正对照pMDC83-GmVAS1-1质粒);B:转基因拟南芥中GmVAS1-1相对表达量
Fig. 8 Identification of transgenic A. thaliana plant A: Screening of transgenic A. thaliana plants(M: DNA marker. 1: Positive control pMDC83-GmVAS1-1 plasmid). B: Relative expression of GmVAS1-1 in transgenic A. thaliana
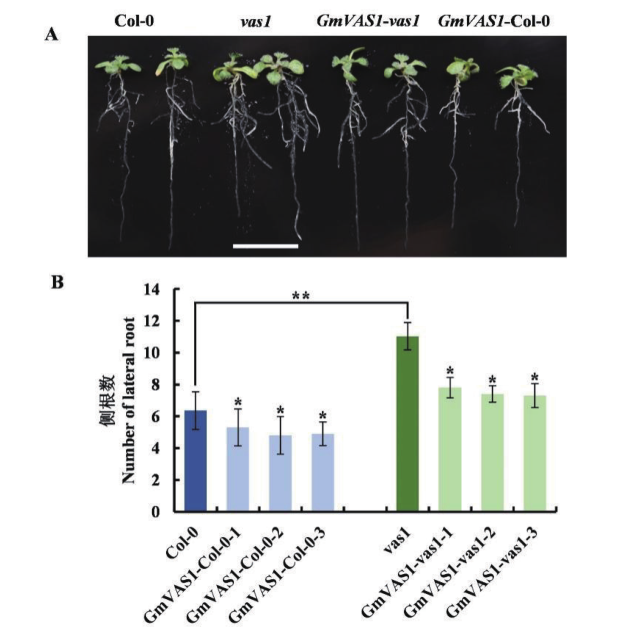
图9 转基因拟南芥侧根发育 A:苗期根部表型;B:侧根数统计
Fig. 9 Lateral root development in transgenic A. thaliana plants A: Phenotype of roots during seedling. B: Statistic of lateral root in various plants
| 基因Gene ID | 基因注释Gene description | 筛选方式Screening method |
|---|---|---|
| Glyma.12G037400 Glyma.14G189400 | 果糖-二磷酸醛缩酶1/果糖-1,6-二磷酸醛缩酶 Fructose-bisphosphate aldolase 1/Fructose-1,6-bisphosphate triosephosphate-lyase | 酵母双杂交试验 Yeast hybrid screening |
| Glyma.04G065600 | 双功能抑制剂/脂质转移蛋白/种子储存2s白蛋白超家族蛋白 Bifunctional inhibitor /Lipid-transfer protein / Seed storage 2s albumin super family protein | |
| Glyma.12G207600 | 成束蛋白样阿拉伯半乳聚糖蛋白13相关蛋白 Fasciclin-like arabinogalactan protein 13-related | |
| Glyma.01G063000 | UDP-阿拉伯吡喃糖突变酶UDP-arabinopyranose mutase | |
| Glyma.06G064900 | 与膜相关的泛素蛋白连接酶//未命名亚族Membrane associated ring finger // Subfamily not named | |
| Glyma.08G261200 Glyma.20G148900 | 甲基四氢叶酸:同型半胱氨酸甲基转移酶相关/同型半胱氨酸S-甲基转移酶 Methyltetrahydrofolate: Homocysteine methyltransferase related/Homocysteine S-methyltransferase | STRING数据库在线预测 STRING database |
| Glyma.02G087900 Glyma.10G172700 Glyma.13G001200 | 蛋氨酸-γ-裂解酶Methionine-gamma-lyase | |
| Glyma.10G244300 Glyma.20G150100 Glyma.20G150200 | 顺式还原酮双加氧酶/1,2-二羟基-3-酮-5-甲硫戊烯双加氧酶4 Acireductone dioxygenase / 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 4 | |
| Glyma.15G012300 | 亚组I转氨酶相关//未命名亚族Subgroup I aminotransferase related // Subfamily not named | |
| Glyma.19G158800 | 同型半胱氨酸S-甲基转移酶/硒代半胱氨酸Se-甲基转移酶 Homocysteine S-methyltransferase/Selenocysteine Se-methyltransferase | |
| Glyma.03G158400 | 吲哚-3-甘油-磷酸裂解酶/组蛋白去乙酰化酶抑制剂/色氨酸合酶/色氨酸合成酶 Indole-3-glycerol-phosphate lyase / TSA / Tryptophan synthase / Tryptophan synthetase |
表2 相互作用因子
Table 2 Interaction factors
| 基因Gene ID | 基因注释Gene description | 筛选方式Screening method |
|---|---|---|
| Glyma.12G037400 Glyma.14G189400 | 果糖-二磷酸醛缩酶1/果糖-1,6-二磷酸醛缩酶 Fructose-bisphosphate aldolase 1/Fructose-1,6-bisphosphate triosephosphate-lyase | 酵母双杂交试验 Yeast hybrid screening |
| Glyma.04G065600 | 双功能抑制剂/脂质转移蛋白/种子储存2s白蛋白超家族蛋白 Bifunctional inhibitor /Lipid-transfer protein / Seed storage 2s albumin super family protein | |
| Glyma.12G207600 | 成束蛋白样阿拉伯半乳聚糖蛋白13相关蛋白 Fasciclin-like arabinogalactan protein 13-related | |
| Glyma.01G063000 | UDP-阿拉伯吡喃糖突变酶UDP-arabinopyranose mutase | |
| Glyma.06G064900 | 与膜相关的泛素蛋白连接酶//未命名亚族Membrane associated ring finger // Subfamily not named | |
| Glyma.08G261200 Glyma.20G148900 | 甲基四氢叶酸:同型半胱氨酸甲基转移酶相关/同型半胱氨酸S-甲基转移酶 Methyltetrahydrofolate: Homocysteine methyltransferase related/Homocysteine S-methyltransferase | STRING数据库在线预测 STRING database |
| Glyma.02G087900 Glyma.10G172700 Glyma.13G001200 | 蛋氨酸-γ-裂解酶Methionine-gamma-lyase | |
| Glyma.10G244300 Glyma.20G150100 Glyma.20G150200 | 顺式还原酮双加氧酶/1,2-二羟基-3-酮-5-甲硫戊烯双加氧酶4 Acireductone dioxygenase / 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 4 | |
| Glyma.15G012300 | 亚组I转氨酶相关//未命名亚族Subgroup I aminotransferase related // Subfamily not named | |
| Glyma.19G158800 | 同型半胱氨酸S-甲基转移酶/硒代半胱氨酸Se-甲基转移酶 Homocysteine S-methyltransferase/Selenocysteine Se-methyltransferase | |
| Glyma.03G158400 | 吲哚-3-甘油-磷酸裂解酶/组蛋白去乙酰化酶抑制剂/色氨酸合酶/色氨酸合成酶 Indole-3-glycerol-phosphate lyase / TSA / Tryptophan synthase / Tryptophan synthetase |
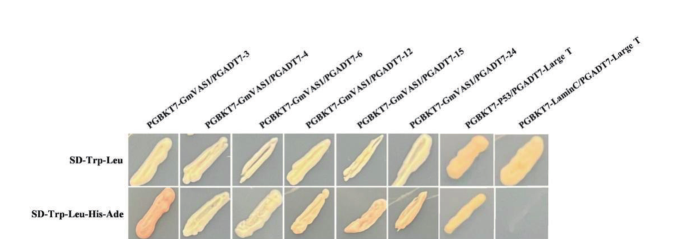
图10 酵母双杂交文库筛选获得的阳性克隆 P53/Large T:系统正对照;Lamin C/Large T:负对照
Fig. 10 Positive clones obtained from yeast two hybrid library screening P53/Large T: The positive control. Lamin C/Large T: The negative control
| [1] |
郭宾会, 戴毅, 宋丽. 干旱下植物激素影响作物根系发育的研究进展[J]. 生物技术通报, 2018, 34(7): 48-56.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0425 |
| Guo BH, Dai Y, Song L. Research progress on the effects of phytohormones on crop root system development under drought condition[J]. Biotechnol Bull, 2018, 34(7): 48-56. | |
| [2] |
Saini S, Sharma I, Kaur N, et al. Auxin: a master regulator in plant root development[J]. Plant Cell Rep, 2013, 32(6): 741-757.
doi: 10.1007/s00299-013-1430-5 pmid: 23553556 |
| [3] | Meng FN, Xiang D, Zhu JS, et al. Molecular mechanisms of root development in rice[J]. Rice(N Y), 2019, 12(1): 1. |
| [4] | Roychoudhry S, Kepinski S. Auxin in root development[J]. Cold Spring Harb Perspect Biol, 2022, 14(4): a039933. |
| [5] |
Zheng ZY, Guo YX, Novák O, et al. Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1[J]. Nat Chem Biol, 2013, 9(4): 244-246.
doi: 10.1038/nchembio.1178 pmid: 23377040 |
| [6] |
Pieck M, et al. Auxin and tryptophan homeostasis are facilitated by the ISS1/VAS1 aromatic aminotransferase in Arabidopsis[J]. Genetics, 2015, 201(1): 185-199.
doi: 10.1534/genetics.115.180356 URL |
| [7] |
Zhang T, Li RN, et al. The YUCCA-auxin-WOX11 module controls crown root development in rice[J]. Front Plant Sci, 2018, 9: 523.
doi: 10.3389/fpls.2018.00523 pmid: 29740464 |
| [8] |
Korver RA, et al. Out of shape during stress: a key role for auxin[J]. Trends Plant Sci, 2018, 23(9): 783-793.
doi: S1360-1385(18)30129-8 pmid: 29914722 |
| [9] |
Kawai T, Akahoshi R, Shelley IJ, et al. Auxin distribution in lateral root primordium development affects the size and lateral root diameter of rice[J]. Front Plant Sci, 2022, 13: 834378.
doi: 10.3389/fpls.2022.834378 URL |
| [10] |
Lv BS, et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis[J]. Mol Plant, 2021, 14(2): 285-297.
doi: 10.1016/j.molp.2020.11.011 pmid: 33221411 |
| [11] |
Krishnamurthy A, Rathinasabapathi B. Auxin and its transport play a role in plant tolerance to arsenite-induced oxidative stress in Arabidopsis thaliana[J]. Plant Cell Environ, 2013, 36(10): 1838-1849.
doi: 10.1111/pce.2013.36.issue-10 URL |
| [12] |
Wang YQ, Chai CL, Valliyodan B, et al. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean(Glycine max)[J]. BMC Genomics, 2015, 16: 951.
doi: 10.1186/s12864-015-2149-1 URL |
| [13] |
Singh VK, Jain M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean[J]. Front Plant Sci, 2015, 6: 918.
doi: 10.3389/fpls.2015.00918 pmid: 26579165 |
| [14] | Zhang Z, Gao L, Ke M, et al. GmPIN1-mediated auxin asymmetry regulates leaf petiole angle and plant architecture in soybean[J]. J Integr Plant Biol, 2022: 2022 Apr 29. |
| [15] |
Tripathi P, Tayade R, Mun BG, et al. Silicon application differentially modulates root morphology and expression of PIN and YUCCA family genes in soybean(Glycine max L.)[J]. Front Plant Sci, 2022, 13: 842832.
doi: 10.3389/fpls.2022.842832 URL |
| [16] |
Hu B, Jin JP, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [17] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [18] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]//The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005: 571-607. |
| [19] |
Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 URL |
| [20] |
Liu SX, Qin B, Fang QX, et al. Genome-wide identification, phylogeny and expression analysis of the bZIP gene family in Alfalfa(Medicago sativa)[J]. Biotechnol Biotechnol Equip, 2021, 35(1): 905-916.
doi: 10.1080/13102818.2021.1938674 URL |
| [21] |
Wu CL, Dai J, Chen ZS, et al. Comprehensive analysis and expression profiles of cassava UDP-glycosyltransferases(UGT)family reveal their involvement in development and stress responses in cassava[J]. Genomics, 2021, 113(5): 3415-3429.
doi: 10.1016/j.ygeno.2021.08.004 URL |
| [22] | 刘薇, 张彦威, 等. 大豆干旱响应GRAS基因筛选及GmGRAS27的生物信息学和逆境表达分析[J]. 大豆科学, 2022, 41(1): 36-42. |
| Liu W, Zhang YW, et al. Screening of soybean drought responsive GRAS genes and bioinformatics and adversity stress expression analysis on GmGRAS27[J]. Soybean Sci, 2022, 41(1): 36-42. | |
| [23] |
Fang YS, Cao D, Yang HL, et al. Genome-wide identification and characterization of soybean GmLOR gene family and expression analysis in response to abiotic stresses[J]. Int J Mol Sci, 2021, 22(22): 12515.
doi: 10.3390/ijms222212515 URL |
| [24] |
Yin LL, Zhang ML, Wu RG, et al. Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response[J]. BMC Plant Biol, 2021, 21(1): 408.
doi: 10.1186/s12870-021-03184-2 |
| [25] |
Gong ZZ, et al. Plant abiotic stress response and nutrient use efficiency[J]. Sci China Life Sci, 2020, 63(5): 635-674.
doi: 10.1007/s11427-020-1683-x pmid: 32246404 |
| [26] |
Le BH, Wagmaister JA, et al. Using genomics to study legume seed development[J]. Plant Physiol, 2007, 144(2): 562-574.
pmid: 17556519 |
| [27] |
Matsuo S, Miyatake K, Endo M, et al. Loss of function of the Pad-1 aminotransferase gene, which is involved in auxin homeostasis, induces parthenocarpy in Solanaceae plants[J]. PNAS, 2020, 117(23): 12784-12790.
doi: 10.1073/pnas.2001211117 URL |
| [28] |
Yoon J, Cho LH, Tun W, et al. Sucrose signaling in higher plants[J]. Plant Sci, 2021, 302: 110703.
doi: 10.1016/j.plantsci.2020.110703 URL |
| [29] | 陈雨生, 江一帆, 张瑛. 中国大豆生产格局变化及其影响因素[J]. 经济地理, 2022, 42(3): 87-94. |
| Chen YS, Jiang YF, Zhang Y. Spatio-temporal evolution and influencing factors of soybean production in China[J]. Econ Geogr, 2022, 42(3): 87-94. | |
| [30] | 付帅. 西南地区玉米-大豆带状复合种植专用晚熟大豆品种的抗旱机制研究[D]. 雅安: 四川农业大学, 2020. |
| Fu S. Drought tolerance mechanism of late-maturing soybean varieties specifically use for maize-soybean strip intercropping in southwest China[D]. Ya'an: Sichuan Agricultural University, 2020. | |
| [31] | 于凤丽, 陈祥金, 于晓光, 等. 大豆抗旱育种研究进展[J]. 黑河学院学报, 2013, 4(1): 124-128. |
| Yu FL, Chen XJ, Yu XG, et al. Research on breeding of soybean drought[J]. J Heihe Univ, 2013, 4(1): 124-128. | |
| [32] |
Fahad S, Bajwa AA, Nazir U, et al. Crop production under drought and heat stress: plant responses and management options[J]. Front Plant Sci, 2017, 8: 1147.
doi: 10.3389/fpls.2017.01147 pmid: 28706531 |
| [33] | 李琬. 干旱对大豆根系生育的影响及灌溉缓解效应研究进展[J]. 草业学报, 2019, 28(4): 192-202. |
| Li W. Research progress in understanding the effects of drought on growth of the soybean root system and the efficiency of irrigation[J]. Acta Prataculturae Sin, 2019, 28(4): 192-202. | |
| [34] |
Ding H, Zhang ZM, Dai LX, et al. Responses of root morphology of peanut varieties differing in drought tolerance to water-deficient stress[J]. Acta Eco Sin, 2013, 33(17): 5169-5176.
doi: 10.5846/stxb URL |
| [1] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [2] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [3] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [4] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [5] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [6] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [7] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [8] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [9] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [10] | 陈奕博, 杨万明, 岳爱琴, 王利祥, 杜维俊, 王敏. 基于SLAF标记的大豆遗传图谱构建及苗期耐盐性QTL定位[J]. 生物技术通报, 2023, 39(2): 70-79. |
| [11] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [12] | 许睿, 祝英方. 中介体复合物在植物非生物胁迫应答中的功能[J]. 生物技术通报, 2023, 39(11): 54-60. |
| [13] | 孙雨桐, 刘德帅, 齐迅, 冯美, 黄栩筝, 姚文孔. 茉莉酸调控植物生长发育和胁迫的研究进展[J]. 生物技术通报, 2023, 39(11): 99-109. |
| [14] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [15] | 安昌, 陆琳, 沈梦千, 陈盛圳, 叶康卓, 秦源, 郑平. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||