








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 82-93.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0337
收稿日期:2025-03-26
出版日期:2025-09-26
发布日期:2025-09-24
作者简介:巩慧玲,女,博士,副教授,研究方向 :植物逆境生理学;E-mail: gonghl@lut.edu.cn基金资助:
GONG Hui-ling( ), XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping
), XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping
Received:2025-03-26
Published:2025-09-26
Online:2025-09-24
摘要:
目的 漆酶(laccase, LACs)是一类含有铜离子的多酚氧化酶,在植物生长发育和胁迫响应过程中发挥重要作用。鉴定马铃薯 StLAC 基因家族成员并分析其表达特征,为 StLAC 功能研究提供参考。 方法 基于马铃薯基因组数据,运用生物信息学方法对马铃薯 StLAC 基因家族进行鉴定;利用转录组数据分析 StLAC 基因在不同组织内和非生物胁迫下的表达模式,并通过 RT-qPCR对其在盐胁迫下表达模式进行验证。 结果 在马铃薯中鉴定到77个 StLAC 基因,其中56个 StLAC 蛋白质等电点大于7,仅 StLAC 69为疏水蛋白,其余76个成员为亲水蛋白。亚细胞定位预测结果表明,20个 StLAC
巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93.
GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses[J]. Biotechnology Bulletin, 2025, 41(9): 82-93.
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| StLAC4 | GTCATTTGGATGTTCACTTGCCTTG | TCTGGTGGAGGAGGAGGTAGC |
| StLAC12 | AGGTCCAGTTGTTTATGCTCAAGAAG | ACACGATAGCCTCTGACGGATG |
| StLAC17 | GGTTGTTCCATTGCCACATTGATTC | CGGTTCCATTGTCTGTTTCTCTCC |
| StLAC26 | AAGGTGATCGTCTTGAAATTGAAGTTG | GTGTTACATAGGCTGGTCCATCTG |
| StLAC36 | ACCTTTGGCACCTCTTTGGAATG | GAGTAGGAGGCTTAGGAGACAACC |
| StLAC40 | TGTCGTTTGTGCCACTTGATACTC | CCTGTATGCTTCCAACTCTGAATCC |
| StLAC43 | ATCGTGTCTGCTACATTGCTCTG | CAAGAGGAGAAGCGGTGATATAAGAG |
| StLAC45 | TGCTATCTTCTCTGCTTCACAACAC | TTCCAATCACCTGCTGCTTAACTC |
| StLAC55 | GGGAAACACCGTTACGCCATC | CACCTTGTCCGCAACACCATAG |
| StLAC56 | AGGCATATAATAGCGTAGTTGGAGTTG | GGGAGGATGGGTGGTGATGAAG |
| StLAC66 | TGTCGTTTGTGCCACTTGATACTC | CCTGTATGCTTCCAACTCTGAATCC |
| StLAC68 | TGCTATCTTCTCTGCTTCACAACAC | TTCCAATCACCTGCTGCTTAACTC |
表1 RT-qPCR引物信息
Table 1 Primer information for RT-qPCR
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| StLAC4 | GTCATTTGGATGTTCACTTGCCTTG | TCTGGTGGAGGAGGAGGTAGC |
| StLAC12 | AGGTCCAGTTGTTTATGCTCAAGAAG | ACACGATAGCCTCTGACGGATG |
| StLAC17 | GGTTGTTCCATTGCCACATTGATTC | CGGTTCCATTGTCTGTTTCTCTCC |
| StLAC26 | AAGGTGATCGTCTTGAAATTGAAGTTG | GTGTTACATAGGCTGGTCCATCTG |
| StLAC36 | ACCTTTGGCACCTCTTTGGAATG | GAGTAGGAGGCTTAGGAGACAACC |
| StLAC40 | TGTCGTTTGTGCCACTTGATACTC | CCTGTATGCTTCCAACTCTGAATCC |
| StLAC43 | ATCGTGTCTGCTACATTGCTCTG | CAAGAGGAGAAGCGGTGATATAAGAG |
| StLAC45 | TGCTATCTTCTCTGCTTCACAACAC | TTCCAATCACCTGCTGCTTAACTC |
| StLAC55 | GGGAAACACCGTTACGCCATC | CACCTTGTCCGCAACACCATAG |
| StLAC56 | AGGCATATAATAGCGTAGTTGGAGTTG | GGGAGGATGGGTGGTGATGAAG |
| StLAC66 | TGTCGTTTGTGCCACTTGATACTC | CCTGTATGCTTCCAACTCTGAATCC |
| StLAC68 | TGCTATCTTCTCTGCTTCACAACAC | TTCCAATCACCTGCTGCTTAACTC |

图7 马铃薯 LAC 基因在不同组织中(A)和不同胁迫下(B)的表达分析
Fig. 7 Expression analysis of LAC gene family of S.tuberosum in different tissues (A) and under different stress (B)
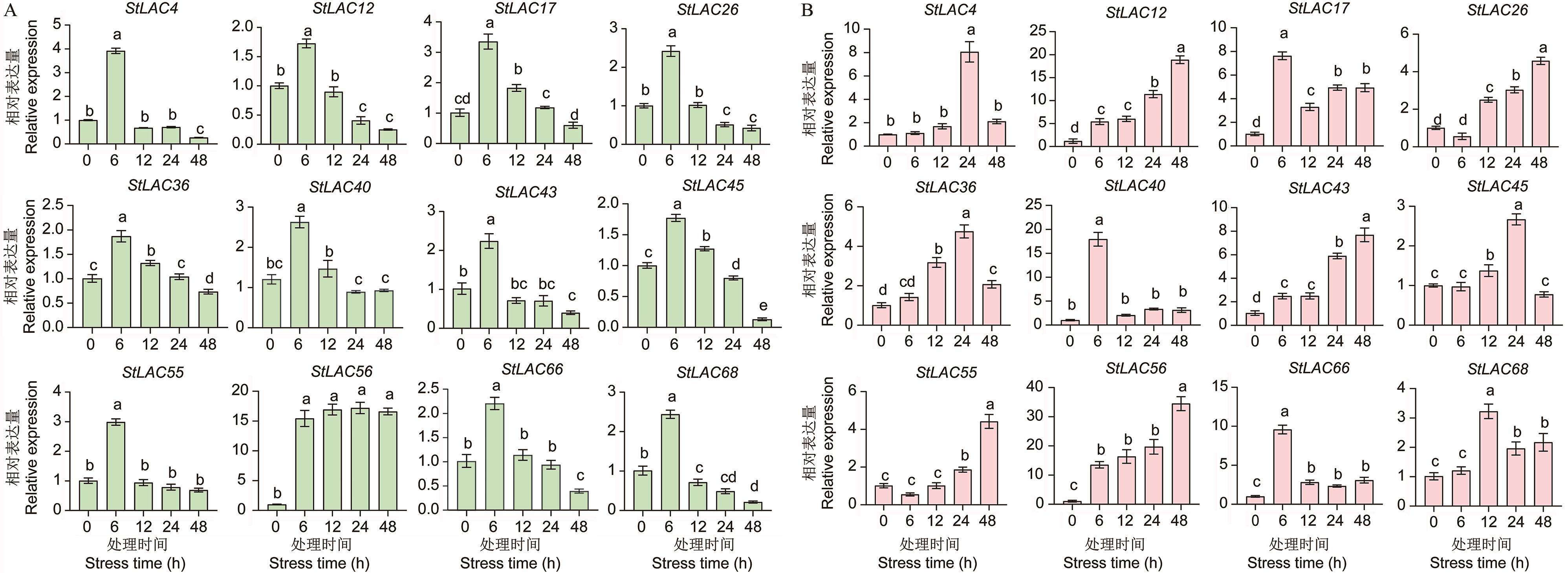
图8 马铃薯根(A)和叶(B)中StLAC基因在盐胁迫下的相对表达量数据为3个独立生物学重复的平均值(± SE),不同字母表示P < 0.05时差异显著
Fig. 8 Relative expressions of StLACs in S. tuberosum roots (A) and leaf (B) under salt stressData are mean (± SE) from three independent biological replicates. Different letters indicate significant differences at P < 0.05
| [1] | 王祎宁, 赵国柱, 谢响明, 等. 漆酶及其应用的研究进展 [J]. 生物技术通报, 2009, 25(5): 35-38. |
| Wang YN, Zhao GZ, Xie XM, et al. Advances in laccase and its applications [J]. Biotechnol Bull, 2009, 25(5): 35-38. | |
| [2] | Yoshida H. Chemistry of lacquer (Urushi) [J]. Journal of the Chemical Society, 1883, 43: 472-486. |
| [3] | Arregui L, Ayala M, Gómez-Gil X, et al. Laccases: structure, function, and potential application in water bioremediation [J]. Microb Cell Fact, 2019, 18(1): 200. |
| [4] | Akram F, Ashraf S, Haq IU, et al. Eminent industrial and biotechnological applications of laccases from bacterial source: a current overview [J]. Appl Biochem Biotechnol, 2022, 194(5): 2336-2356. |
| [5] | McCaig BC, Meagher RB, Dean JFD. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana [J]. Planta, 2005, 221(5): 619-636. |
| [6] | 张盛春, 鞠常亮, 王小菁. 拟南芥漆酶基因AtLAC4参与生长及非生物胁迫响应 [J]. 植物学报, 2012, 47(4): 357-365. |
| Zhang SC, Ju CL, Wang XJ. Arabidopsis laccase gene AtLAC4 regulates plant growth and responses to abiotic stress [J]. Chin Bull Bot, 2012, 47(4): 357-365. | |
| [7] | Ranocha P, Chabannes M, Chamayou S, et al. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar [J]. Plant Physiol, 2002, 129(1): 145-155. |
| [8] | Liu QQ, Luo L, Wang XX, et al. Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis [J]. Int J Mol Sci, 2017, 18(2): 209. |
| [9] | Gu HL, Wang Y, Xie H, et al. Drought stress triggers proteomic changes involving lignin, flavonoids and fatty acids in tea plants [J]. Sci Rep, 2020, 10(1): 15504. |
| [10] | Liu MY, Dong H, Wang M, et al. Evolutionary divergence of function and expression of laccase genes in plants [J]. J Genet, 2020, 99: 23. |
| [11] | Berthet S, Demont-Caulet N, Pollet B, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems [J]. Plant Cell, 2011, 23(3): 1124-1137. |
| [12] | Hu Q, Min L, Yang XY, et al. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis [J]. Plant Physiol, 2018, 176(2): 1808-1823. |
| [13] | 王国栋. 过量表达棉花分泌型漆酶拟南芥对酚酸类物质和三氯苯酚的植物体外修复 [D]. 上海: 中国科学院研究生院(上海生命科学研究院), 2004. |
| Wang GD. Phytoremediation ex planta of phenolics and trichlorophenol by overexpressing a cotton secretory laccase in Arabidopsis [D]. Shanghai: Shanghai Institutes for Biological Sciences, 2004. | |
| [14] | 陈宇航. OsLAC12调控水稻耐热性功能分析 [D]. 荆州: 长江大学, 2025. |
| Chen YH. Function analysis of OsLAC12 regulation of heat tolerance of rice [D]. Jingzhou: Yangtze University, 2025. | |
| [15] | Wei JZ, Tirajoh A, Effendy J, et al. Characterization of salt-induced changes in gene expression in tomato (Lycopersicon esculentum) roots and the role played by abscisic acid [J]. Plant Sci, 2000, 159(1): 135-148. |
| [16] | Liang MX, Haroldsen V, Cai XN, et al. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress [J]. Plant Cell Environ, 2006, 29(5): 746-753. |
| [17] | 徐小勇, 顾铭洲, 梁梦鸽, 等. 枳漆酶基因家族鉴定及其响应盐胁迫的表达分析 [J]. 江苏农业科学, 2023, 51(9): 52-59. |
| Xu XY, Gu MZ, Liang MG, et al. Genome-wide identification of the LAC gene family and its expression analysis under salt stress in Poncirus trifoliata [J]. Jiangsu Agric Sci, 2023, 51(9): 52-59. | |
| [18] | Cai XN, Davis EJ, Ballif J, et al. Mutant identification and characterization of the laccase gene family in Arabidopsis [J]. J Exp Bot, 2006, 57(11): 2563-2569. |
| [19] | 吕昕培. 梭梭木质素合成对盐和渗透胁迫的响应及HaLAC15和HaCOMT的功能鉴定 [D]. 兰州: 兰州大学, 2023. |
| Lyu XP. Responses of lignin synthesis in Haloxylon ammodendron to salt and osmotic stresses and functional identification of HaLAC15 and HaCOMT [D]. Lanzhou: Lanzhou University, 2023. | |
| [20] | Cho HY, Lee CH, Hwang SG, et al. Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis [J]. Gene, 2014, 552(1): 98-105. |
| [21] | Ma J, Xu ZS, Wang F, et al. Isolation, purification and characterization of two laccases from carrot (Daucus carota L.) and their response to abiotic and metal ions stresses [J]. Protein J, 2015, 34(6): 444-452. |
| [22] | Visser RGF, Bachem CWB, de Boer JM, et al. Sequencing the potato genome: outline and first results to come from the elucidation of the sequence of the world’s third most important food crop [J]. Am J Potato Res, 2009, 86(6): 417-429. |
| [23] | 张琨, 袁利兵, 彭志红, 等. 马铃薯转基因工程研究现状 [J]. 湖南农业科学, 2011(5): 6-8, 11. |
| Zhang K, Yuan LB, Peng ZH, et al. Research status quo of potato transgenic engineering [J]. Hunan Agric Sci, 2011(5): 6-8, 11. | |
| [24] | Bernards MA, Fleming WD, Llewellyn DB, et al. Biochemical characterization of the suberization-associated anionic peroxidase of potato [J]. Plant Physiol, 1999, 121(1): 135-146. |
| [25] | Ha JH, Moon KB, Kim MS, et al. The laccase promoter of potato confers strong Tuber-specific expression in transgenic plants [J]. Plant Cell Tissue Organ Cult, 2015, 120(1): 57-68. |
| [26] | Machinandiarena MF, Oyarburo NS, Daleo GR, et al. The reinforcement of potato cell wall as part of the phosphite-induced tolerance to UV-B radiation [J]. Biologia Plant, 2018, 62(2): 388-394. |
| [27] | Zheng XY, Zhang XJ, Zhao JM, et al. Meyerozyma guilliermondii promoted the deposition of GSH type lignin by activating the biosynthesis and polymerization of monolignols at the wounds of potato tubers [J]. Food Chem, 2023, 416: 135688. |
| [28] | Wang Q, Li G, Zheng KJ, et al. The soybean laccase gene family: evolution and possible roles in plant defense and stem strength selection [J]. Genes, 2019, 10(9): 701. |
| [29] | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server [J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| [30] | 杨阳, 刘媛媛. 苹果MdLAC基因家族成员的鉴定与序列分析 [J]. 河南农业科学, 2021, 50(7): 125-135. |
| Yang Y, Liu YY. Identification and sequence analysis of the MdLAC gene family members in apple [J]. J Henan Agric Sci, 2021, 50(7): 125-135. | |
| [31] | 周一鹏. 柑橘漆酶基因家族的鉴定及抗逆性研究 [D]. 扬州: 扬州大学, 2018. |
| Zhou YP. Identification and stress resistance of Citrus laccase gene family [D]. Yangzhou: Yangzhou University, 2018. | |
| [32] | 白永生, 周嘉杰, 唐玉林. 植物漆酶及其在植物生长发育中的作用 [J]. 生命科学, 2022, 34(9): 1135-1144. |
| Bai YS, Zhou JJ, Tang YL. Plant laccases and their roles in plant growth and development [J]. Chin Bull Life Sci, 2022, 34(9): 1135-1144. | |
| [33] | 刘清泉. 铜胁迫下水稻木质素合成的响应机制及水稻漆酶在植物重金属耐性中的作用 [D]. 南京: 南京农业大学, 2015. |
| Liu QQ. Response mechanism of lignin synthesis in rice under copper stress and the role of rice laccase in plants tolerance to heavy metal [D]. Nanjing: Nanjing Agricultural University, 2015. | |
| [34] | Le Roy J, Blervacq AS, Créach A, et al. Spatial regulation of monolignol biosynthesis and laccase genes control developmental and stress-related lignin in flax [J]. BMC Plant Biol, 2017, 17(1): 124. |
| [35] | 薛佳丽, 任晓庆, 郄倩茹, 等. 谷子LAC基因家族的鉴定及分析 [J]. 山西农业科学, 2021, 49(4): 395-402. |
| Xue JL, Ren XQ, Qie QR, et al. Identification and analysis of LAC gene family in millet [J]. J Shanxi Agric Sci, 2021, 49(4): 395-402. | |
| [36] | 姚文, 刘苗苗, 陈沫池, 等. 苋菜LAC基因家族成员全基因组鉴定与表达分析 [J]. 中国蔬菜, 2024,(11). 48-56. |
| Yao W, Liu MM, Cheng MC, et al. Genome-wide identification and expression analysis of LAC gene family of Amaranthus tricolor L. [J]. Chinese Vegetable, 2024,(11). 48-56. | |
| [37] | 李郑华, 姜成东, 王鹏, 等. 荔枝漆酶(LAC)基因家族鉴定及表达分析 [J]. 分子植物育种, 2023, 21(7): 2174-2187. |
| Li ZH, Jiang CD, Wang P, et al. Identification and expression analysis of laccase(LAC) genes in Litchi (Litchi chinensis Sonn.) [J]. Mol Plant Breed, 2023, 21(7): 2174-2187. | |
| [38] | Birchler JA, Yang H. The multiple fates of gene duplications: deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation [J]. Plant Cell, 2022, 34(7): 2466-2474. |
| [39] | 邱可立, 王玉民, 何金铃, 等. 桃漆酶家族基因鉴定及PpLAC21功能分析 [J]. 园艺学报, 2022, 49(6): 1351-1362. |
| Qiu KL, Wang YM, He JL, et al. Identification of peach laccase family genes and functional analysis of PpLAC21 [J]. Journal of Horticulture, 2022, 49(6): 1351-1362. | |
| [40] | Salvi HM, Yadav GD. Process intensification using immobilized enzymes for the development of white biotechnology [J]. Catal Sci Technol, 2021, 11(6): 1994-2020. |
| [1] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [2] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [3] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [4] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [5] | 化文平, 刘菲, 浩佳欣, 陈尘. 丹参ADH基因家族的鉴定与表达模式分析[J]. 生物技术通报, 2025, 41(8): 211-219. |
| [6] | 腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233. |
| [7] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [8] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [9] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [10] | 李雅琼, 格桑拉毛, 陈启迪, 杨宇环, 何花转, 赵耀飞. 异源过表达高粱SbSnRK2.1增强拟南芥对盐胁迫的抗性[J]. 生物技术通报, 2025, 41(8): 115-123. |
| [11] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [12] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [13] | 张泽, 杨秀丽, 宁东贤. 花生4CL基因家族鉴定及对干旱与盐胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 117-127. |
| [14] | 付博晗, 毛华, 赵薪程, 陆虹, 欧庸彬, 姚银安. 不同杨树SOS1基因启动子的克隆及盐胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 205-213. |
| [15] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||