








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 128-138.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1255
李凯月1( ), 邓晓霞1, 殷缘1, 杜亚彤1, 徐元静1, 王竞红1, 于耸2, 蔺吉祥1(
), 邓晓霞1, 殷缘1, 杜亚彤1, 徐元静1, 王竞红1, 于耸2, 蔺吉祥1( )
)
收稿日期:2024-12-24
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
蔺吉祥,男,教授,博士生导师,研究方向 :植物逆境生理生态学;E-mail: linjixiang@nefu.edu.cn作者简介:李凯月,女,硕士研究生,研究方向 :蓖麻耐铝机制;E-mail: 2023120534@nefu.edu.cn
基金资助:
LI Kai-yue1( ), DENG Xiao-xia1, YIN Yuan1, DU Ya-tong1, XU Yuan-jing1, WANG Jing-hong1, YU Song2, LIN Ji-xiang1(
), DENG Xiao-xia1, YIN Yuan1, DU Ya-tong1, XU Yuan-jing1, WANG Jing-hong1, YU Song2, LIN Ji-xiang1( )
)
Received:2024-12-24
Published:2025-07-26
Online:2025-07-22
摘要:
目的 从蓖麻全基因组范围内鉴定RcLEA基因家族成员,分析其基因特征和潜在功能,为揭示LEA在调控蓖麻铝胁迫耐受性中的作用奠定基础。 方法 利用生物信息学方法对蓖麻LEA家族基因进行鉴定,并对其理化性质、系统进化关系、基因结构、保守基序、启动子顺式作用元件,以及该家族部分成员在铝胁迫处理后的表达情况进行分析。 结果 从蓖麻基因组中共鉴定到52个LEA家族基因成员,根据系统发育分析,其可分为RcLEA_1、RcLEA_2、RcLEA_3、RcLEA_4、RcLEA_5、RcLEA_6、Rc_DHN和Rc_SMP 8个亚族,分布于10条染色体上;家族成员氨基酸数量在91‒406 aa之间;等电点介于4.54‒10.29之间;分子量在9 951.42‒44 515.78 Da之间;大多数LEA蛋白为亲水性蛋白;基序分析显示不同亚家族之间的motif有很大差异,但在同一亚类中是相似的。基因结构分析显示,多数基因有1个或1个以上的内含子;52个基因启动子区域均存在1种或多种与植物激素和逆境响应相关的顺式作用元件;共线性分析显示,鉴定到8对复制基因。另外,实时荧光定量PCR结果显示,RcLEA基因在铝胁迫处理下,与对照相比基因表达量发生变化,推测该家族基因参与铝胁迫应答。 结论 从蓖麻基因组中共鉴定出52个RcLEA基因,分为8个亚族。对8个亚族部分RcLEA基因进行铝胁迫表达分析,发现大部分基因表达量变化显著,表明这些基因在蓖麻响应铝胁迫中发挥重要作用。
李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138.
LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L.[J]. Biotechnology Bulletin, 2025, 41(7): 128-138.
| 基因 Gene | 上游引物 Forward primer (5′‒3′) | 下游引物 Reverse primer (5′‒3′) |
|---|---|---|
RcLEA1 RcLEA16 | AAGGAGCGTAGGAAGGCAAGAG ACCATGCGATTGAATGCCACTA | GTGGTGGTGTCTCGACTTCTGA TGTGTAGCTGTCCACCGTCAA |
| RcLEA24 | CCTTCCATGTCGGCACAGATG | CCAAACCAACAACTGTCCCTCT |
| RcLEA36 | TGCGTCCTGTAAACCCTAATGC | ATTACCACCACTACCAGCAGCA |
| RcLEA38 | ATGGCTCGCTCTCACTCCTAC | AGTCGCCGAATAGCCTCTCAG |
| RcLEA40 | TGGGTCTGTGCTGAAGAAAGTTG | TCTCATTTACAACATCGTGTGG |
RcLEA41 RcLEA43 | GGTGGACGTTGCATATGCAGATC TACGATAGAGAGAAAGGAGGAT | TCCGCCCTTCAGCAAGGTGTTC ATAGTAGAAACATCAGCCTCCC |
| RcLEA45 | TGGTGGGAGAAGGAAGAAAGGA | ATGACCGTGTTGTTCTGCTGAA |
| RcLEA50 | GGTTGGTGCGGAGGTTGTTG | CCAGCAGCAGAGTAAGCAGTTG |
| ACTIN | TCCCTCAGTACGTTCCAGCA | CACCTCCATACTCCTCCCT |
表1 实时荧光定量PCR引物及碱基序列
Table 1 Real-time fluorescent quantitative PCR primers and base sequences
| 基因 Gene | 上游引物 Forward primer (5′‒3′) | 下游引物 Reverse primer (5′‒3′) |
|---|---|---|
RcLEA1 RcLEA16 | AAGGAGCGTAGGAAGGCAAGAG ACCATGCGATTGAATGCCACTA | GTGGTGGTGTCTCGACTTCTGA TGTGTAGCTGTCCACCGTCAA |
| RcLEA24 | CCTTCCATGTCGGCACAGATG | CCAAACCAACAACTGTCCCTCT |
| RcLEA36 | TGCGTCCTGTAAACCCTAATGC | ATTACCACCACTACCAGCAGCA |
| RcLEA38 | ATGGCTCGCTCTCACTCCTAC | AGTCGCCGAATAGCCTCTCAG |
| RcLEA40 | TGGGTCTGTGCTGAAGAAAGTTG | TCTCATTTACAACATCGTGTGG |
RcLEA41 RcLEA43 | GGTGGACGTTGCATATGCAGATC TACGATAGAGAGAAAGGAGGAT | TCCGCCCTTCAGCAAGGTGTTC ATAGTAGAAACATCAGCCTCCC |
| RcLEA45 | TGGTGGGAGAAGGAAGAAAGGA | ATGACCGTGTTGTTCTGCTGAA |
| RcLEA50 | GGTTGGTGCGGAGGTTGTTG | CCAGCAGCAGAGTAAGCAGTTG |
| ACTIN | TCCCTCAGTACGTTCCAGCA | CACCTCCATACTCCTCCCT |
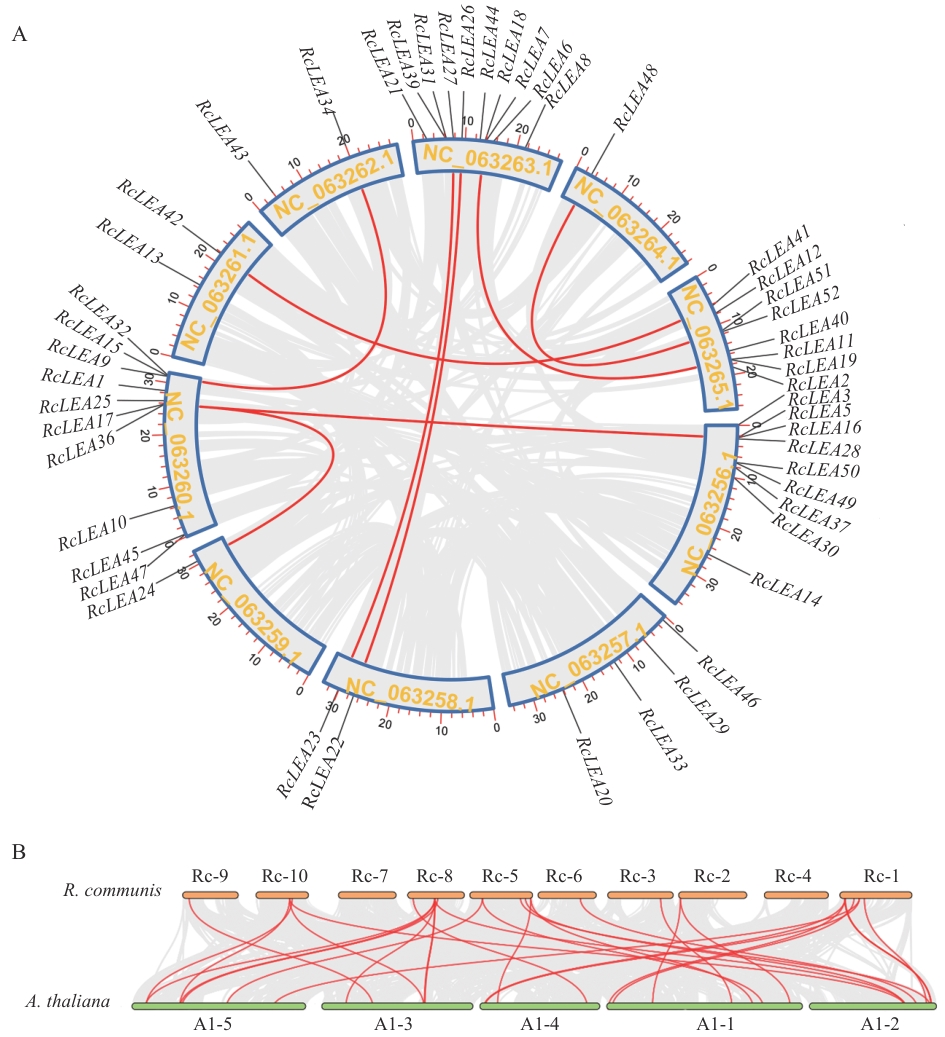
图5 蓖麻种内RcLEA基因共线性分析(A)和蓖麻与拟南芥LEA基因同源性分析(B)
Fig. 5 Synteny analysis of RcLEA genes within Ricinus species (A) and homology analysis of LEA genes between R. communis and Arabidopsis (B)

图6 蓖麻LEA基因在铝胁迫处理下的表达量分析不同小写字母表示差异显著(P<0.05)
Fig. 6 Expression analysis of R. communis LEA genes under aluminum stressDifferent lowercase letters indicate significant differences (P<0.05)
| [1] | Salazar-Chavarría V, Sánchez-Nieto S, Cruz-Ortega R. Fagopyrum esculentum at early stages copes with aluminum toxicity by increasing ABA levels and antioxidant system [J]. Plant Physiol Biochem, 2020, 152: 170-176. |
| [2] | Chandra J, Keshavkant S. Mechanisms underlying the phytotoxicity and genotoxicity of aluminum and their alleviation strategies: a review [J]. Chemosphere, 2021, 278: 130384. |
| [3] | Wang Y, Yao ZS, Zhan Y, et al. Potential benefits of liming to acid soils on climate change mitigation and food security [J]. Glob Chang Biol, 2021, 27(12): 2807-2821. |
| [4] | 冯晶. 水稻耐铝候选基因生物学功能分析 [D]. 沈阳: 沈阳农业大学, 2023. |
| Feng J. Biological function analysis of aluminum-tolerant candidate gene in rice [D]. Shenyang: Shenyang Agricultural University, 2023. | |
| [5] | 黄丽媛, 胡光伟, 王琼, 等. 铝胁迫下钙对油茶营养元素吸收积累的影响 [J]. 湖北农业科学, 2022, 61(22): 19-24. |
| Huang LY, Hu GW, Wang Q, et al. Effect of calcium on nutrient uptake and accumulation in Camellia oleifera under aluminum stress [J]. Hubei Agric Sci, 2022, 61(22): 19-24. | |
| [6] | 周小华, 李昆志, 赵峥, 等. 外源抗坏血酸对水稻抗铝生理指标的影响 [J]. 热带作物学报, 2021, 42(3): 769-776. |
| Zhou XH, Li KZ, Zhao Z, et al. Effects of exogenous ascorbic acid on physiological indexes in rice under aluminum stress [J]. Chin J Trop Crops, 2021, 42(3): 769-776. | |
| [7] | 刘爱慧. BnaALMT7.2在甘蓝型油菜根系响应铝毒害的作用机制研究 [D]. 重庆: 西南大学, 2021. |
| Liu AH. Study on the mechanism of BnaALMT7.2 in the root system of Brassica napus in response to aluminum toxicity [D]. Chongqing: Southwest University, 2021. | |
| [8] | Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana [J]. BMC Genomics, 2008, 9: 118. |
| [9] | Xiao BZ, Huang YM, Tang N, et al. Over-expression of a LEA gene in rice improves drought resistance under the field conditions [J]. Theor Appl Genet, 2007, 115(1): 35-46. |
| [10] | Zan T, Li LQ, Li JT, et al. Genome-wide identification and characterization of late embryogenesis abundant protein-encoding gene family in wheat: Evolution and expression profiles during development and stress [J]. Gene, 2020, 736: 144422. |
| [11] | Liu Y, Wang L, Xing X, et al. ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses [J]. Plant Cell Physiol, 2013, 54(6): 944-959. |
| [12] | Hand SC, Menze MA, Toner M, et al. LEA proteins during water stress: not just for plants anymore [J]. Annu Rev Physiol, 2011, 73: 115-134. |
| [13] | Stacy RA, Aalen RB. Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis-abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis [J]. Planta, 1998, 206(3): 476-478. |
| [14] | Close TJ, Lammers PJ. An osmotic stress protein of cyanobacteria is immunologically related to plant dehydrins [J]. Plant Physiol, 1993, 101(3): 773-779. |
| [15] | Abba' S, Ghignone S, Bonfante P. A dehydration-inducible gene in the truffle Tuber borchii identifies a novel group of dehydrins [J]. BMC Genomics, 2006, 7: 39. |
| [16] | Iii LD. A repeating 11-mer amino acid motif and plant desiccation [J]. Plant J, 1993, 3(3): 363-369. |
| [17] | Punta M, Coggill PC, Eberhardt RY, et al. The pfam protein families database [J]. Nucleic Acids Res, 2012, 40(database issue): D290-D301. |
| [18] | 杨洪强, 贾文锁, 张大鹏. 植物水分胁迫信号识别与转导 [J]. 植物生理学通讯, 2001, 37(2): 149-154. |
| Yang HQ, Jia WS, Zhang DP. Plant signal perception and transduction response to water stress [J]. Plant Physiol Commun, 2001, 37(2): 149-154. | |
| [19] | Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response [J]. Plant Physiol, 1997, 115(2): 327-334. |
| [20] | 李佳琪. 沙地云杉LEA基因家族鉴定及PmLEA25基因抗逆功能分析 [D]. 呼和浩特: 内蒙古农业大学, 2022. |
| Li JQ. Identification of LEA gene family of Picea mongolica and analysis of stress resistance function of PmLEA25 gene [D]. Hohhot: Inner Mongolia Agricultural University, 2022. | |
| [21] | 刘星辰. 紫花苜蓿MsLEA2基因的克隆及其功能研究 [D]. 上海: 上海交通大学, 2019. |
| Liu XC. Cloning and functional analysis of MsLEA2 gene in Alfalfa (Medicago sativa Linn) [D]. Shanghai: Shanghai Jiao Tong University, 2019. | |
| [22] | 刘洋. 玉米LEA蛋白基因ZmDHN13,ZmLEA3和ZmLEA5C的分离与功能分析[D]. 泰安: 山东农业大学, 2014. |
| Liu Y. Isolation and functional analysis of LEA protein genes, ZmDHN13, ZmLEA3 and ZmLEA5C in Zea mays [D]. Tai'an: Shandong Agricultural University, 2014. | |
| [23] | 黄若兰. 花生AhLecRK9和AhLEAs在铝胁迫下的功能研究 [D]. 南宁: 广西大学, 2022. |
| Huang RL. Functional analysis of AhLecRK9 and AhLEAs in peanut under aluminum stress [D]. Nanning: Guangxi University, 2022. | |
| [24] | Yu AM, Li F, Liu AZ. Comparative proteomic and transcriptomic analyses provide new insight into the formation of seed size in castor bean [J]. BMC Plant Biol, 2020, 20(1): 48. |
| [25] | Scarpa A, Guerci A. Various uses of the castor oil plant (Ricinus communis L.). A review [J]. J Ethnopharmacol, 1982, 5(2): 117-137. |
| [26] | Thanh NC, Narayanan M, Saravanan M, et al. Bio/phyremediation potential of Leptospirillum ferrooxidans and Ricinus communis on metal contaminated mine sludge [J]. Chemosphere, 2023, 339: 139739. |
| [27] | 张涛, 吴心悦, 闫靖芸, 等. 蓖麻SAP基因家族的鉴定及生物信息学分析 [J]. 黑龙江农业科学, 2025(3): 12-21. |
| Zhang T, Wu XY, Yan JY, et al. Identification and bioinformatics analysis of SAP gene family in castor [J]. Heilongjiang Agric Sci, 2025(3): 12-21. | |
| [28] | 王冬艳, 张春玲, 冯研, 等. 蓖麻bZIP基因家族的全基因组鉴定、生物信息学及表达分析 [J]. 安徽农学通报, 2025, 31(2): 27-35. |
| Wang DY, Zhang CL, Feng Y, et al. Genome-wide identification, bioinformatics and expression analysis of bZIP gene family of Ricinus communis [J]. Anhui Agric Sci Bull, 2025, 31(2): 27-35. | |
| [29] | 张童劼, 孙一, 杨跃超, 等. 蓖麻CNGC基因家族的鉴定与系统进化分析 [J]. 内蒙古民族大学学报: 自然科学版, 2024, 39(6): 70-75. |
| Zhang TJ, Sun Y, Yang YC, et al. Identification and phylogenetic analysis of Ricinus communis L. CNGC gene family [J]. J Inn Mong Minzu Univ Nat Sci Ed, 2024, 39(6): 70-75. | |
| [30] | 李艳肖, 向殿军, 满丽莉, 等. 蓖麻NAC基因家族鉴定及表达分析 [J]. 农业生物技术学报, 2023, 31(12): 2519-2534. |
| Li YX, Xiang DJ, Man LL, et al. Identification and expression analysis of NAC gene family in castor(Ricinus communis) [J]. J Agric Biotechnol, 2023, 31(12): 2519-2534. | |
| [31] | 邹智, 黄启星, 安锋. 蓖麻LEA基因家族的全基因组鉴定、分类及其进化分析 [J]. 中国油料作物学报, 2013, 35(6): 637-643. |
| Zou Z, Huang QX, An F. Genome-wide identification, classification and phylogenetic analysis of LEA gene family in castor bean (Ricinus communis L.) [J]. Chin J Oil Crop Sci, 2013, 35(6): 637-643. | |
| [32] | Lu JJ, Pan C, Fan W, et al. A chromosome-level genome assembly of wild castor provides new insights into its adaptive evolution in tropical desert [J]. Genomics Proteomics Bioinformatics, 2022, 20(1): 42-59. |
| [33] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [34] | 刘浩. 小麦LEA基因家族的鉴定及脱水蛋白WZY2响应干旱胁迫的调节机制 [D]. 杨凌: 西北农林科技大学, 2021. |
| Liu H. Genome-wide identification of late embryogenesis abundant (LEA) gene family and regulatory mechanism of dehydrin WZY2 in response to drought stress in wheat [D]. Yangling: Northwest A & F University, 2021. | |
| [35] | Candat A, Paszkiewicz G, Neveu M, et al. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress [J]. Plant Cell, 2014, 26(7): 3148-3166. |
| [36] | 李震. 胡麻LEA基因家族的鉴定及其在种子发育过程中的遗传效应 [D]. 北京: 中国农业科学院, 2021. |
| Li Z. Identification and genetic effect of LEA genes during seed development in linseed flax [D]. Beijing: Chinese Academy of Agricultural Sciences, 2021. | |
| [37] | 王琪. 人参LEA基因家族鉴定分析与PgLEA2-50应答非生物胁迫研究 [D]. 长春: 吉林农业大学, 2024. |
| Wang Q. Identification and analysis of LEA gene family in Panax ginseng and the role of PgLEA2-50 in abiotic stress [D]. Changchun: Jilin Agricultural University, 2024. | |
| [38] | 陈凯, 佟晓楠, 张晓媛, 等. 枳LEA基因家族鉴定及其对非生物胁迫的响应 [J]. 西北植物学报, 2023, 43(6): 918-928. |
| Chen K, Tong XN, Zhang XY, et al. Genome-wide identification and abiotic stress responses of LEA gene family in Poncirus trifoliata [J]. Acta Bot Boreali Occidentalia Sin, 2023, 43(6): 918-928. | |
| [39] | 于晓云. 月季LEA基因家族鉴定与弯刺蔷薇低温响应的脱水素基因发掘 [D]. 福州: 福建农林大学, 2022. |
| Yu XY. Genome-wide identification of LEA gene family in Rosa chinensis and the mining of cold-response dehydrin genes in R . beggeriana [D]. Fuzhou: Fujian Agriculture and Forestry University, 2022. | |
| [40] | 杨剑. 谷子LEA_2基因家族鉴定及SiLEA14参与谷子逆境胁迫响应的功能研究 [D]. 太原: 山西大学, 2023. |
| Yang J. Identification of SiLEA_2 gene family and functional analysis of SiLEA14 in Setaria italica stress response [D]. Taiyuan: Shanxi University, 2023. | |
| [41] | 贾金山. 顽拗性三七种子LEA基因家族的鉴定与表达分析 [D]. 昆明: 云南农业大学, 2023. |
| Jia JS. Identification and expression analysis of the LEA gene family in recalcitrant Panax notoginseng seeds [D]. Kunming: Yunnan Agricultural University, 2023. | |
| [42] | 唐晓飞, 刘鑫磊, 薛永国, 等. 大豆LEA-2的家族基因鉴定与表达特征分析 [J/OL]. 分子植物育种, 2023. . |
| Tang XF, Liu XL, Xue YG, et al. Genome-wide identification and expression analysis of LEA-2 gene family in soybean [J/OL]. Mol Plant Breed, 2023. . | |
| [43] | 付娆, 李佳佳, 单燕, 等. 栽培种花生LEA蛋白基因家族的鉴定及其低温胁迫下表达分析 [J]. 花生学报, 2022, 51(3): 1-12. |
| Fu R, Li JJ, Shan Y, et al. Genome-wide identification and expression analysis of the late embryogenesis abundant (LEA) protein gene family in cultivated peanut (Arachis hypogaea L.) on its response to cold stress [J]. J Peanut Sci, 2022, 51(3): 1-12. | |
| [44] | 周璇. 垫状卷柏LEA1基因的结构与功能分析 [D]. 昆明: 西南林业大学, 2022. |
| Zhou X. Structural and functional analysis of the LEA1 gene in Selaginella pulvinata [D]. Kunming: Southwest Forestry University, 2022. | |
| [45] | Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor [J]. Trends Genet, 2008, 24(8): 375-378. |
| [46] | Liang Y, Xiong ZY, Zheng JX, et al. Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus [J]. Sci Rep, 2016, 6: 24265. |
| [47] | Wu CL, Hu W, Yan Y, et al. The late embryogenesis abundant protein family in cassava (Manihot esculenta crantz): genome-wide characterization and expression during abiotic stress [J]. Molecules, 2018, 23(5): 1196. |
| [48] | 耿伟博. 烟草胚胎发育晚期丰富蛋白(LEA蛋白)基因家族对非生物胁迫反应的鉴定与分析 [D]. 泰安: 山东农业大学, 2022. |
| Geng WB. Genome-wide identification and analysis of the tobacco late embryogenesis abundant proteins (LEA proteins) gene family in response to abiotic stresses [D]. Tai'an: Shandong Agricultural University, 2022. | |
| [49] | Sachs M. Alteration of gene expression during environmental stress in plants [J]. Annu Rev Plant Physiol Plant Mol Biol, 37: 363-376. |
| [50] | Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins [J]. Science, 1989, 245(4916): 371-378. |
| [1] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [2] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [3] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [4] | 王苗苗, 赵相龙, 王召明, 刘志鹏, 闫龙凤. 花苜蓿TCP基因家族的鉴定及其在干旱胁迫下的表达模式分析[J]. 生物技术通报, 2025, 41(6): 179-190. |
| [5] | 瞿美玲, 周思敏, 张惊宇, 何佳蔚, 朱佳源, 刘笑蓉, 童巧珍, 周日宝, 刘湘丹. 灰毡毛忍冬bHLH转录基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(6): 256-268. |
| [6] | 刘鑫, 王嘉雯, 李进伟, 牟策, 杨盼盼, 明军, 徐雷锋. 兰州百合三个LdBBXs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(5): 186-196. |
| [7] | 赵婧, 郭茜, 李睿琦, 雷滢炀, 岳爱琴, 赵晋忠, 殷丛丛, 杜维俊, 牛景萍. 大豆GmGST基因簇基因序列分析及诱导表达分析[J]. 生物技术通报, 2025, 41(5): 129-140. |
| [8] | 罗嗣芳, 张祖铭, 谢丽芳, 郭紫晶, 陈兆星, 杨月华, 严翔, 张洪铭. 山金柑GATA基因家族全基因组鉴定及在果实发育中的表达分析[J]. 生物技术通报, 2025, 41(5): 218-230. |
| [9] | 李志强, 罗正乾, 徐琳黎, 周国慧, 屈丝雨, 刘恩良, 顼东婷. 基于T2T基因组鉴定大豆R2R3-MYB基因家族及干旱和盐胁迫下的表达分析[J]. 生物技术通报, 2025, 41(5): 141-152. |
| [10] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| [11] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [12] | 孙天国, 衣兰, 秦旭洋, 乔梦雪, 谷新颖, 韩艺, 沙伟, 张梅娟, 马天意. 大白菜DABB基因家族的全基因组鉴定及盐碱胁迫下的表达分析[J]. 生物技术通报, 2025, 41(4): 156-165. |
| [13] | 王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175. |
| [14] | 王天禧, 杨炳松, 潘荣君, 盖文贤, 梁美霞. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究[J]. 生物技术通报, 2025, 41(4): 176-187. |
| [15] | 黄金恒, 黄茜, 张家燕, 周新裕, 廖沛然, 杨全. 广金钱草C3H基因家族鉴定及不同品种表达分析[J]. 生物技术通报, 2025, 41(4): 243-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||