








生物技术通报 ›› 2021, Vol. 37 ›› Issue (6): 13-23.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1343
郝向阳1( ), 刘范1, 武欢1, 王斌1, 孙雪丽1, 项蕾蕾1, 王天池1, 赖钟雄1(
), 刘范1, 武欢1, 王斌1, 孙雪丽1, 项蕾蕾1, 王天池1, 赖钟雄1( ), 程春振1,2(
), 程春振1,2( )
)
收稿日期:2020-11-02
出版日期:2021-06-26
发布日期:2021-07-08
作者简介:郝向阳,女,硕士,研究方向:花卉生物技术;E-mail: 基金资助:
HAO Xiang-yang1( ), LIU Fan1, WU Huan1, WANG Bin1, SUN Xue-li1, XIANG Lei-lei1, WANG Tian-chi1, LAI Zhong-xiong1(
), LIU Fan1, WU Huan1, WANG Bin1, SUN Xue-li1, XIANG Lei-lei1, WANG Tian-chi1, LAI Zhong-xiong1( ), CHENG Chun-zhen1,2(
), CHENG Chun-zhen1,2( )
)
Received:2020-11-02
Published:2021-06-26
Online:2021-07-08
摘要:
为了解非洲菊PAL基因编码蛋白的理化性质和表达模式,以非洲菊品种“玲珑”为材料,利用RACE和RT-PCR技术克隆获得4个PAL基因(命名为GjPAL1-GjPAL4),利用生物信息学方法研究它们的结构及其编码蛋白的特性。利用qPCR检测GjPALs在不同组织部位和不同逆境胁迫处理下的表达模式。序列分析显示:4个GjPAL的CDS为2 115-2 136 bp,编码蛋白长度为704-711 aa,均无信号肽的稳定酸性亲水蛋白。实时荧光定量PCR结果显示,GjPAL1在叶中表达量最高,GjPAL2-GjPAL4均在根中表达量最高;4个GjPALs的表达量均受低温显著抑制;GjPAL1和GjPAL2的表达受SA、NaCl和PEG显著抑制;GjPAL3和GjPAL4在SA处理下呈先升后降的表达趋势,GjPAL3的表达受NaCl和PEG的显著诱导;隐地疫霉处理条件下,GjPAL1和GjPAL4的表达量显著升高,GjPAL2的表达量显著降低,而GjPAL3的表达量呈先降后升趋势。GjPALs广泛参与非洲菊对不同逆境的应答过程,各成员在不同逆境处理下的作用存在一定差异。
郝向阳, 刘范, 武欢, 王斌, 孙雪丽, 项蕾蕾, 王天池, 赖钟雄, 程春振. 非洲菊GjPAL的克隆及表达分析[J]. 生物技术通报, 2021, 37(6): 13-23.
HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni[J]. Biotechnology Bulletin, 2021, 37(6): 13-23.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Application |
|---|---|---|---|
| GjPAL1F-outer | GGATTACGGGTTCAAAGGT | 3' RACE | |
| GjPAL1F-inner | GCTCCAGTTTCTTGCTAATCC | ||
| GjPAL4F-outer | TAAGCCAAGTAGCCAAGAAGGT | ||
| GjPAL4F-inner | AGGACTTGCTTCGTGTGGTT | ||
| GjPAL1R-outer | CGGCGTTCAAGAATCTGA | 5'RACE | |
| GjPAL1R-inner | GTGACACCGTAACTATCAGTCCC | ||
| GjPAL1F | GAGTCCAGTGTGTGAAAC | 2 301 | RT-PCR |
| GjPAL1R | AATGGTGAGCCTCTTCCGA | ||
| GjPAL2F | TGGATCATACCAATGGAAATGAC | 2 137 | |
| GjPAL2R | CACTTATGAAATGGGAAGAGGG | ||
| GjPAL3F | ATGGACAGTAAAGCCGTCAAGATC | 2 204 | |
| GjPAL3R | CTAACAAATTGGAAGAGGAACAC | ||
| GjPAL4F | GAGTCTAGTGTGTGAAACTTTGATAGC | 2 117 | |
| GjPAL4R | TTAACATATCGGAAGAGGCTCAC | ||
| GjPAL1-qF | TTCCGAACAGAATCAAGGC | 125 | qRT-PCR |
| GjPAL1-qR | TGACCCTTACACATTGCCG | ||
| GjPAL2-qF | TGCATGAATAGTGATCCATTG | 145 | |
| GjPAL2-qR | GATTCTGACAACTCCACCTGT | ||
| GjPAL3-qF | TCACATAACCAGCCATTTCG | 173 | |
| GjPAL3-qR | ACCATCCGCTTCACTTCGT | ||
| GjPAL4-qF | CGATAATCAATGGAGAACGG | 172 | |
| GjPAL4-qR | TCCACCATCTGTTTCACCTC | ||
| 18sF | TCAAAGCAAGCCTACGCTCT | 125 | |
| 18sR | GCTTTCGCAGTTGTTCGTCT |
表1 基因克隆和定量引物信息
Table 1 Information of the primers used for gene cloning and qRT-PCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Application |
|---|---|---|---|
| GjPAL1F-outer | GGATTACGGGTTCAAAGGT | 3' RACE | |
| GjPAL1F-inner | GCTCCAGTTTCTTGCTAATCC | ||
| GjPAL4F-outer | TAAGCCAAGTAGCCAAGAAGGT | ||
| GjPAL4F-inner | AGGACTTGCTTCGTGTGGTT | ||
| GjPAL1R-outer | CGGCGTTCAAGAATCTGA | 5'RACE | |
| GjPAL1R-inner | GTGACACCGTAACTATCAGTCCC | ||
| GjPAL1F | GAGTCCAGTGTGTGAAAC | 2 301 | RT-PCR |
| GjPAL1R | AATGGTGAGCCTCTTCCGA | ||
| GjPAL2F | TGGATCATACCAATGGAAATGAC | 2 137 | |
| GjPAL2R | CACTTATGAAATGGGAAGAGGG | ||
| GjPAL3F | ATGGACAGTAAAGCCGTCAAGATC | 2 204 | |
| GjPAL3R | CTAACAAATTGGAAGAGGAACAC | ||
| GjPAL4F | GAGTCTAGTGTGTGAAACTTTGATAGC | 2 117 | |
| GjPAL4R | TTAACATATCGGAAGAGGCTCAC | ||
| GjPAL1-qF | TTCCGAACAGAATCAAGGC | 125 | qRT-PCR |
| GjPAL1-qR | TGACCCTTACACATTGCCG | ||
| GjPAL2-qF | TGCATGAATAGTGATCCATTG | 145 | |
| GjPAL2-qR | GATTCTGACAACTCCACCTGT | ||
| GjPAL3-qF | TCACATAACCAGCCATTTCG | 173 | |
| GjPAL3-qR | ACCATCCGCTTCACTTCGT | ||
| GjPAL4-qF | CGATAATCAATGGAGAACGG | 172 | |
| GjPAL4-qR | TCCACCATCTGTTTCACCTC | ||
| 18sF | TCAAAGCAAGCCTACGCTCT | 125 | |
| 18sR | GCTTTCGCAGTTGTTCGTCT |
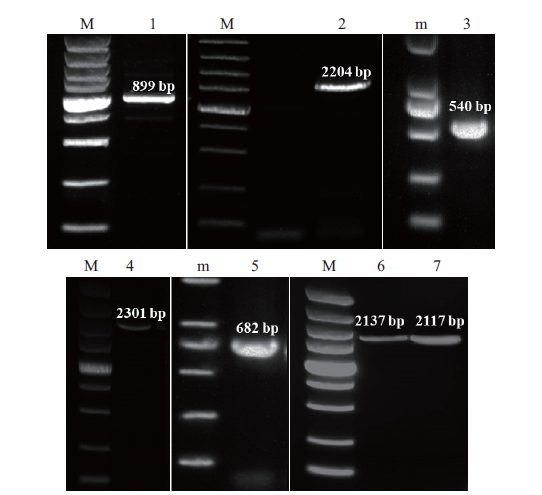
图1 PCR产物电泳图 1-7分别为:GjPAL1 3'RACE、GjPAL3 RT-PCR、GjPAL1 5'RACE、GjPAL1 RT-PCR、GjPAL4 3'RACE、GjPAL2 RT-PCR和GjPAL4 RT-PCR验证结果;M:DL5000 DNA marker(条带从下到上依次为:100 bp、250 bp、500 bp、750 bp、1 000 bp、1 500 bp、2 000 bp、3 000 bp和5 000 bp);m:DL2000 DNA marker(条带从下到上依次为:100 bp、250 bp、500 bp、750 bp、1 000 bp和2 000 bp)
Fig. 1 Electrophoresis result of PCR products 1-7 is the electrophoresis result of GjPAL1 3' RACE, GjPAL3 RT-PCR, GjPAL1 5' RACE, GjPAL1 RT-PCR, GjPAL4 3' RACE, GjPAL2 RT-PCR and GjPAL4 RT-PCR products, respectively. M: DL5000 DNA marker (the band from bottom to top is 100 bp, 250 bp, 500 bp, 750 bp, 1 000 bp, 1 500 bp, 2 000 bp, 3 000 bp and 5 000 bp, respectively). m: DL2000 DNA marker (the band from bottom to top is 100 bp, 250 bp, 500 bp, 750 bp, 1000 bp and 2000 bp, respectively)
| 蛋白 Protein | 分子量 Molecular weight/kD | 分子式 Formula | 等电点pI | 脂肪系数 Aliphatic index | 不稳定系数 Instability index | 总亲水性 GRAVY |
|---|---|---|---|---|---|---|
| GjPAL1 | 77 052.27 | C3405H5452N938O1033S31 | 6.18 | 90.49 | 31.79 | -0.134 |
| GjPAL2 | 76 538.09 | C3384H5389N925O1044S25 | 5.53 | 91.97 | 34.20 | -0.125 |
| GjPAL3 | 77 326.50 | C3428H5470N940O1036S28 | 5.93 | 92.07 | 34.24 | -0.091 |
| GjPAL4 | 77 091.13 | C3406H5453N941O1038S28 | 6.18 | 91.05 | 32.98 | -0.152 |
表2 4个GjPALs蛋白基本理化性质分析结果
Table 2 Physicochemical properties of the four GjPALs
| 蛋白 Protein | 分子量 Molecular weight/kD | 分子式 Formula | 等电点pI | 脂肪系数 Aliphatic index | 不稳定系数 Instability index | 总亲水性 GRAVY |
|---|---|---|---|---|---|---|
| GjPAL1 | 77 052.27 | C3405H5452N938O1033S31 | 6.18 | 90.49 | 31.79 | -0.134 |
| GjPAL2 | 76 538.09 | C3384H5389N925O1044S25 | 5.53 | 91.97 | 34.20 | -0.125 |
| GjPAL3 | 77 326.50 | C3428H5470N940O1036S28 | 5.93 | 92.07 | 34.24 | -0.091 |
| GjPAL4 | 77 091.13 | C3406H5453N941O1038S28 | 6.18 | 91.05 | 32.98 | -0.152 |

图3 不同植物PAL氨基酸序列的多重序列比对 黑框代表酶活性中心序列,红框代表典型的保守区域
Fig. 3 Multiple alignment result of PALs from different plant species Characters in the black box represent the enzyme activity center and the red box represent the typical conserved region of the PAL enzyme activity center
| 蛋白 Protein | 长度 Length/aa | 卷曲螺旋结构 Coiled coil structure | 跨膜结构域Transmembrane domain | 磷酸化修饰位点Phosphorylation site | |||||
|---|---|---|---|---|---|---|---|---|---|
| 外→内Outside→Inside | 内→外Inside→Outside | S | T | Y | |||||
| GjPAL1 | 708 | 无 | 3 | 5 | 33 | 21 | 7 | ||
| GjPAL2 | 704 | 有 | 3 | 7 | 34 | 20 | 7 | ||
| GjPAL3 | 711 | 有 | 5 | 6 | 30 | 24 | 8 | ||
| GjPAL4 | 708 | 无 | 2 | 5 | 36 | 19 | 8 | ||
表3 GjPAL跨膜结构域及磷酸化修饰位点分析结果
Table 3 Result of transmembrane structure and phospho-rylation sites in GjPALs
| 蛋白 Protein | 长度 Length/aa | 卷曲螺旋结构 Coiled coil structure | 跨膜结构域Transmembrane domain | 磷酸化修饰位点Phosphorylation site | |||||
|---|---|---|---|---|---|---|---|---|---|
| 外→内Outside→Inside | 内→外Inside→Outside | S | T | Y | |||||
| GjPAL1 | 708 | 无 | 3 | 5 | 33 | 21 | 7 | ||
| GjPAL2 | 704 | 有 | 3 | 7 | 34 | 20 | 7 | ||
| GjPAL3 | 711 | 有 | 5 | 6 | 30 | 24 | 8 | ||
| GjPAL4 | 708 | 无 | 2 | 5 | 36 | 19 | 8 | ||
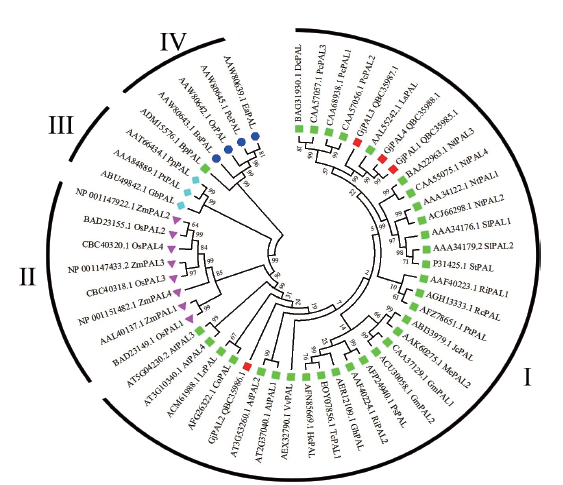
图5 GjPALs和其他植物PALs系统发育树 I:双子叶植物;II:单子叶植物;III:裸子植物;IV:蕨类植物
Fig. 5 Phylogenetic tree for GjPALs and PALs from some other plant species I: Dicotyledons. II: Monocotyledons. III: Gymnosperm. IV: Pteridophyte

图6 GjPAL基因在不同组织部位和不同逆境处理下的表达情况 A:不同组织器官;B:SA处理;C:NaCl处理;D:PEG处理;E:低温处理;F:隐地疫霉接种;*表示0.05水平上显著差异;**表示0.01水平上极显著差异
Fig.6 Relative expression of GjPAL genes in different tissues and organs and under different stress treatments A: Different organs. B: SA treatment. C: NaCl treatment. D: PEG treatment. E: Cold stress treatment. F: Phytophthora cryptogea innoculation treatment. “*” indicates significant difference at the 0.05 level, and “**” indicates very significant difference at the 0.01 level
| [1] |
Costa MA, Collins RE, Anterola AM, et al. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof[J]. Phytochemistry, 2003, 64(6):1097-1112.
doi: 10.1016/S0031-9422(03)00517-X URL |
| [2] |
Huang JL, Gu M, Lai ZB, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress[J]. Plant Physiology, 2010, 153(4):1526-1538.
doi: 10.1104/pp.110.157370 URL |
| [3] |
Ferrer JL, Austin MB, Stewart Jr C, et al. Structure and function of enzymes involved in the biosynjournal of phenylpropanoids[J]. Plant Physiology and Biochemistry, 2008, 46(3):356-370.
doi: 10.1016/j.plaphy.2007.12.009 pmid: 18272377 |
| [4] |
Zhang XB, Liu CJ. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynjournal of phenylpropanoids[J]. Molecular Plant, 2015, 8:17-27.
doi: 10.1016/j.molp.2014.11.001 URL |
| [5] |
Jun SY, Sattler SA, Cortez GS, et al. Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase[J]. Plant Physiology, 2018, 176(2):1452-1468.
doi: 10.1104/pp.17.01608 URL |
| [6] |
De Jong F, Hanley SJ, Beale MH, et al. Characterisation of the willow phenylalanine ammonia-lyase(PAL)gene family reveals expression differences compared with poplar[J]. Phytochemistry, 2015, 117:90-97.
doi: 10.1016/j.phytochem.2015.06.005 URL |
| [7] | 郝向阳, 孙雪丽, 王天池, 等. 植物PAL基因及其编码蛋白的特征与功能研究进展[J]. 热带作物学报, 2018, 39(7):1452-1461. |
| Hao XY, Sun XL, Wang TC, et al. Characteristics and functions of plant Phenylalanine Ammonia Lyase genes and the encoded proteins[J]. Chinese Journal of Tropical Crops, 2018, 39(7):1452-1461. | |
| [8] |
Cochrane FC, Davin LB, Lewis NG. The Arabidopsis phenylalanine ammonia lyase gene family:kinetic characterization of the four PAL isoforms[J]. Phytochemistry, 2004, 65(11):1557-1564.
pmid: 15276452 |
| [9] | 曾嘉丽, 欧阳林娟, 刘家林, 等. 水稻PAL基因的全基因组分析及胁迫表达研究[J]. 基因组学与应用生物学, 2018, 37(9):3881-3888. |
| Ceng JL, Ouyang LJ, Liu JL, et al. Whole genome analysis and stress expression research of PAL gene in rice[J]. Genomics and Applied Biology, 2018, 37(9):3881-3888. | |
| [10] |
Tsai CJ, Harding SA, Tschaplinski TJ, et al. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus[J]. New phytologist, 2006, 172:47-62.
doi: 10.1111/nph.2006.172.issue-1 URL |
| [11] | Gho YS, Kim SJ, Jung KH. Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice[J]. Genes & genomics, 2020, 42:67-76. |
| [12] |
Chandrasekaran M, Belachew ST, Yoon E, et al. Expression of β-1, 3-glucanase(GLU)and phenylalanine ammonia-lyase(PAL)genes and their enzymes in tomato plants induced after treatment with Bacillus subtilis CBR05 against Xanthomonas campestris pv. vesicatoria[J]. Journal of General Plant Pathology, 2017, 83:7-13.
doi: 10.1007/s10327-016-0692-5 URL |
| [13] | 熊飞, 卢秦华, 房婉萍, 等. 基于全基因组的茶树PAL家族基因鉴定及其在生物与非生物胁迫下的表达分析[J]. 园艺学报, 2020, 47(3):517-528. |
| Xiong F, Lu QH, Fang WP, et al. Genome-wide Identification and expression analyses of PAL genes under biotic and abiotic stress in Camellia sinensis[J]. Acta Horticulturae Sinica, 2020, 47(3):517-528. | |
| [14] | 杨郁文, 李双, 黄俊宇, 等. 陆地棉苯丙氨酸解氨酶家族基因的鉴定及分析[J]. 分子植物育种, 2017, 15(4):1184-1191. |
| Yang YW, Li S, Huang JY, et al. Identification and analysis of the gene family of phenylalanine ammonialyase in upland cotton[J]. Molecular Plant Breeding, 2017, 15(4):1184-1191. | |
| [15] |
Nugroho LH, Verberne MC, Verpoorte R. Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants[J]. Plant Physiology and Biochemistry, 2002, 40(9):755-760.
doi: 10.1016/S0981-9428(02)01437-7 URL |
| [16] |
Chaman ME, Copaja SV, Argandoña VH. Relationships between salicylic acid content, phenylalanine ammonia-lyase(PAL)activity, and resistance of barley to aphid infestation[J]. Journal of Agricultural and Food Chemistry, 2003, 51(8):2227-2231.
doi: 10.1021/jf020953b URL |
| [17] |
Ogawa D, Nakajima N, Seo S, et al. The phenylalanine pathway is the main route of salicylic acid biosynjournal in Tobacco mosaic virus-infected tobacco leaves[J]. Plant Biotechnology, 2006, 23(4):395-398.
doi: 10.5511/plantbiotechnology.23.395 URL |
| [18] |
Kim DS, Hwang BK. An important role of the pepper phenylalanine ammonia-lyase gene(PAL1)in salicylic acid-dependent signalling of the defence response to microbial pathogens[J]. Journal of Experimental Botany, 2014, 65(9):2295-2306.
doi: 10.1093/jxb/eru109 URL |
| [19] | 孟祥春, 彭建宗, 王小菁. 光和糖对非洲菊花色素苷积累及CHS、DFR基因表达的影响[J]. 园艺学报, 2007, 34(1):227-230. |
| Meng XC, Peng JZ, Wang XJ. Anthocyanin accumulation and CHS, DFR gene expression regulated by light and sugar in Gerbera hybrid ray floret[J]. Acta Horticulturae Sinica, 2007, 34(1):227-230. | |
| [20] | 王晰, 徐哲, 赖齐贤, 等. 非洲菊切花弯茎影响因素研究进展[J]. 园艺学报, 2015, 42(9):1771-1780. |
| Wang X, Xu Z, Lai QX, et al. Research progress ininfluence factors of stem bending of cut gerbera flower[J]. Acta Horticulturae Sinica, 2015, 42(9):1771-1780. | |
| [21] |
Zhong CM, Tang Y, Pang B, et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida[J]. Horticulture Research, 2020, 7:78.
doi: 10.1038/s41438-020-0296-2 URL |
| [22] | 赖齐贤, 包志毅, 朱祝军, 等. 干旱胁迫对转基因(PSAG12-ipt)非洲菊光合作用的影响[J]. 园艺学报, 2007, 34(1):157-162. |
| Lai QX, Bao ZY, Zhu ZJ, et al. Effects of drought stress on photosynjournal of gerbera modified by PSAG12-ipt[J]. Acta Horticulturae Sinica, 2007, 34(1):157-162. | |
| [23] | 刘芳, 高原, 张竞颐, 等. 非洲菊白粉病病原鉴定及蜡蚧轮枝菌防治试验[J]. 园艺学报, 2010, 37(11):1803-1810. |
| Liu F, Gao Y, Zhang JY, et al. Pathogen identification of gerbera powdery mildew and its control experiment with Verticillium lecanii[J]. Acta Horticulturae Sinica, 2010, 37(11):1803-1810. | |
| [24] | 郝向阳, 林觅, 孙雪丽, 等. 福建非洲菊产区根腐病病原菌的分离与鉴定[J]. 福建农业学报, 2018, 33(4):391-395. |
| Hao XY, Lin M, Sun XL, et al. Isolation and identification of pathogen of gerbera root rot disease in Fujian[J]. Fujian Journal of Agricultural Sciences, 2018, 33(4):391-395. | |
| [25] | Munir N, Cheng CZ, Xia CS, et al. RNA-Seq analysis reveals an essential role of tyrosine metabolism pathway in response to root-rot infection in Gerbera hybrida[J]. PLoS One, 2019, 14(10):e223519. |
| [26] | 郝向阳, 孙雪丽, 刘范, 等. 非洲菊4个POD基因的克隆及表达分析[J]. 西北植物学报, 2018, 38(10):1777-1786. |
| Hao XY, Sun XL, Liu F, et al. Cloning and expression analysis of four gerbera peroxidase genes[J]. Acta Botanica Boreali-Occidentalia Sinica, 2018, 38(10):1777-1786. | |
| [27] |
Ritter H, Schulz GE. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase[J]. The Plant Cell, 2004, 16(12):3426-3436.
doi: 10.1105/tpc.104.025288 URL |
| [28] |
Wang Z, Li JY, Jia CH, et al. Molecular cloning and expression of four phenylalanine ammonia lyase genes from banana interacting with Fusarium oxysporum[J]. Biologia Plantarum, 2016, 60(3):459-468.
doi: 10.1007/s10535-016-0619-1 URL |
| [29] | 许锋, 朱俊, 张风霞, 等. 国槐苯丙氨酸解氨酶基因的克隆、反义表达载体构建及遗传转化[J]. 林业科学研究, 2008, 21(5):611-618. |
| Xu F, Zhu J, Zhang FX, et al. Cloning of PAL gene from Sophora japonica, construction of anti-sense gene of SjPAL and its genetic transformation in Arabidopsis[J]. Forest Research, 2008, 21(5):611-618. | |
| [30] | 许锋, 曹腾, 宁迎晶, 等. 夏枯草苯丙氨酸解氨酶基因的克隆与表达分析[J]. 华北农学报, 2012, 27(1):39-44. |
| Xu F, Cao T, Ning YJ, et al. Molecular cloning and expression analysis of a phenylalanne ammonial-lyase gene from Prunella vulgaris[J]. Acta Agriculturae Boreali-Sinica, 2012, 27(1):39-44. | |
| [31] |
Song J, Wang ZZ. Molecular cloning, expression and characterization of a phenylalanine ammonia-lyase gene(SmPAL1)from Salvia miltiorrhiza[J]. Molecular Biology Reports, 2009, 36(5):939-952.
doi: 10.1007/s11033-008-9266-8 URL |
| [32] |
Shang QM, Li L, Dong CJ. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L.[J]. Planta, 2012, 236(4):1093-1105.
doi: 10.1007/s00425-012-1659-1 URL |
| [33] | 孙梓健, 汤青林, 宋明, 等. 红叶芥低温胁迫下苯丙氨酸解氨酶活性及其基因的克隆表达[J]. 西南大学学报:自然科学版, 2010, 32(2):90-94. |
| Sun ZJ, Tang QL, Song M, et al. Cloning and expression of PAL gene and PAL activity assay in red-leaf mustard(Brassica juncea var. garrhiza Tsen et Lee)under low temperature stress[J]. Journal of Southwest University:Natural Science Edition, 2010, 32(2):90-94. | |
| [34] |
Morkunas I, Bednarski W, Kopyra M. Defense strategies of pea embryo axes with different levels of sucrose to Fusarium oxysporum and Ascochyta pisi[J]. Physiological and Molecular Plant Pathology, 2008, 72(4):167-178.
doi: 10.1016/j.pmpp.2008.09.003 URL |
| [35] |
Shadle GL, Wesley SV, Korth KL, et al. Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase[J]. Phytochemistry, 2003, 64:153-161.
pmid: 12946414 |
| [36] |
Chen YP, Li FJ, Tian L, et al. The phenylalanine ammonia lyase gene LjPAL1 is involved in plant defense responses to pathogens and plays diverse roles in Lotus japonicus-rhizobium symbioses[J]. Molecular Plant-Microbe Interactions, 2017, 30(9):739-753.
doi: 10.1094/MPMI-04-17-0080-R URL |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [4] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [5] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [6] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [7] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [8] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [9] | 侯瑞泽, 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰. 普通白菜PRR5的克隆、表达及功能验证[J]. 生物技术通报, 2023, 39(10): 128-135. |
| [10] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [11] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [12] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| [13] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [14] | 陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61. |
| [15] | 杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||