








生物技术通报 ›› 2022, Vol. 38 ›› Issue (6): 147-156.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1174
收稿日期:2021-09-13
出版日期:2022-06-26
发布日期:2022-07-11
作者简介:杨佳宝,男,硕士研究生,研究方向:植物油脂代谢调控;E-mail: 基金资助:
YANG Jia-bao1( ), ZHOU Zhi-ming1, ZHANG Zhan2, FENG Li1, SUN Li1(
), ZHOU Zhi-ming1, ZHANG Zhan2, FENG Li1, SUN Li1( )
)
Received:2021-09-13
Published:2022-06-26
Online:2022-07-11
摘要:
探究长链酰基辅酶A合成酶(long chain acyl-CoA synthetase,LACS)基因在向日葵(Helianthus annuus L.)油脂积累和逆境响应中的功能,为其在向日葵油脂合成和抗逆中的应用奠定基础。通过RT-PCR克隆得到向日葵HaLACS1的CDS序列,运用生物信息学方法分析HaLACS1的特点。利用实时荧光定量PCR(qRT-PCR)技术检测HaLACS1的组织表达特性及对NaCl、PEG和ABA的响应情况。通过构建GFP和HaLACS1的融合表达载体,转化拟南芥原生质体进行亚细胞定位分析。将HaLACS1转入酿酒酵母突变型菌株YB525中进行功能互补试验,并进行底物偏好性分析。结果显示,HaLACS1开放阅读框1 980 bp,编码659个氨基酸。蛋白进化树分析表明,HaLACS1与拟南芥AtLACS1和莴苣(Lactuca sativa)LsLACS1具有较高的相似性。拟南芥原生质体瞬时表达分析显示HaLACS1定位于内质网。实时荧光定量PCR结果显示,HaLACS1在所有组织中均有表达,但在种子发育的早期表达量较高,花次之。NaCl、PEG和ABA处理均能诱导HaLACS1在向日葵根、茎和叶中的表达。酵母功能互补试验证明HaLACS1蛋白具有LACS酶活性。底物偏好性分析表明HaLACS1偏好棕榈酸(C16:0)和油酸(C18:1)。表明HaLACS1与向日葵种子发育过程中的油脂积累和非生物胁迫响应相关。
杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156.
YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae[J]. Biotechnology Bulletin, 2022, 38(6): 147-156.
| 引物名称 Primer | 引物序列 Primer sequence(5'-3') | 功能 Function |
|---|---|---|
| L1F | CCCAAGCTTATGGATAAGCTG- CAGTTTGC | HaLACS1克隆及酵母表达载体构建 |
| L1R | CCGCTCGAGTTAAGACTTGTT- CCCGCCTA | |
| qF | GAGCTCGAAAACGGAACGTG | qRT-PCR |
| qR | ATTGCAAGCCTCCATCGCTA | |
| PF | CGGACTAGTATGGATAAGCT- GCAGTTT | pAN580-HaLACS1-eGFP载体构建 |
| PR | CATGCCATGGAGACTTGTTC- CCGCCTA | |
| 18S-F | CTACCACATCCAAGGAAGG- CAG | qRT-PCR |
| 18S-R | CGACAGAAGGGACGAGTA- AACC |
表1 引物信息
Table 1 Information of primers used in this study
| 引物名称 Primer | 引物序列 Primer sequence(5'-3') | 功能 Function |
|---|---|---|
| L1F | CCCAAGCTTATGGATAAGCTG- CAGTTTGC | HaLACS1克隆及酵母表达载体构建 |
| L1R | CCGCTCGAGTTAAGACTTGTT- CCCGCCTA | |
| qF | GAGCTCGAAAACGGAACGTG | qRT-PCR |
| qR | ATTGCAAGCCTCCATCGCTA | |
| PF | CGGACTAGTATGGATAAGCT- GCAGTTT | pAN580-HaLACS1-eGFP载体构建 |
| PR | CATGCCATGGAGACTTGTTC- CCGCCTA | |
| 18S-F | CTACCACATCCAAGGAAGG- CAG | qRT-PCR |
| 18S-R | CGACAGAAGGGACGAGTA- AACC |

图2 不同植物LACS1蛋白序列多重比对结果 At:拟南芥;Ha:向日葵;Ls:莴苣;Bn:甘蓝型油菜;Zm:玉米;Os:水稻
Fig. 2 Multiple alignment results of LACS1 proteins from different plants At:Arabidopsis thaliana. Ha:H. annuus L. Ls:Lactuca sativa. Bn:Brassica napus. Zm:Zea mays. Os:Oryza sativa
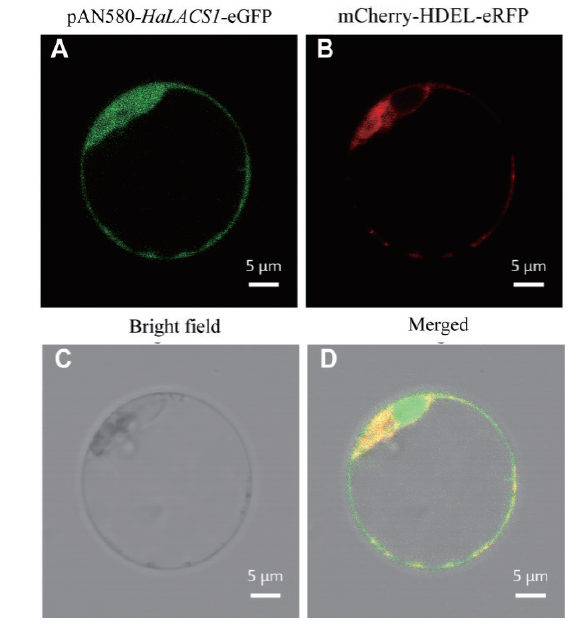
图5 HaLACS1在拟南芥原生质体中的亚细胞定位 A:HaLACS1的GFP(绿色)荧光;B:mCherry-HDEL(内质网marker)的红色荧光;C:明场;D:融合荧光
Fig. 5 Subcellular localization of the HaLACS1 protein in the protoplasts of Arabidopsis A:The green fluorescent signals of the HaLACS1 gene. B:Endoplasmic reticulum marker. C:Bright field. D:Merged
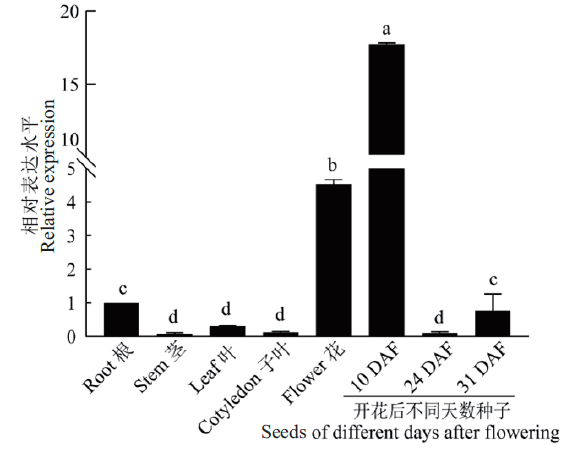
图6 HaLACS1在不同组织中的表达 不同字母表示数值有显著差异(P<0.05)。下同
Fig. 6 Tissue expression analysis of the HaLACS1 gene Different letters indicate values are significantly different(P<0.05). The same below
| [1] | 马惠茹, 等. 内蒙古河套地区向日葵饲料资源生产情况及开发利用现状[J]. 中国畜牧兽医, 2014, 41(3):251-254. |
| Ma HR, et al. Development and production of sunflower by-product feed resource in Hetao area in Inner Mongolia[J]. China Animal Husb Vet Med, 2014, 41(3):251-254. | |
| [2] | 汪磊, 谭美莲, 傅春玲, 等. 利用近红外技术预测向日葵籽仁品质性状[J]. 中国油料作物学报, 2020, 42(1):147-153. |
| Wang L, Tan ML, Fu CL, et al. Prediction of qualitative characteristics of sunflower husked seed by near infrared spectroscopy[J]. Chin J Oil Crop Sci, 2020, 42(1):147-153. | |
| [3] | Hryvusevich P, Navaselsky I, Talkachova Y, et al. Sodium influx and potassium efflux currents in sunflower root cells under high salinity[J]. Front Plant Sci, 2020, 11:613936. |
| [4] |
Kelly AA, Quettier AL, Shaw E, et al. Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis[J]. Plant Physiol, 2011, 157(2):866-875.
doi: 10.1104/pp.111.181784 URL |
| [5] |
Ding LN, Gu SL, Zhu FG, et al. Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus[J]. BMC Plant Biol, 2020, 20(1):21.
doi: 10.1186/s12870-020-2240-x URL |
| [6] |
Groot PH, et al. Fatty acid activation:specificity, localization, and function[J]. Adv Lipid Res, 1976, 14:75-126.
pmid: 3952 |
| [7] |
Hayashi H, De Bellis L, Hayashi Y, et al. Molecular characterization of an Arabidopsis acyl-coenzyme a synthetase localized on glyoxysomal membranes[J]. Plant Physiol, 2002, 130(4):2019-2026.
doi: 10.1104/pp.012955 URL |
| [8] |
Pei ZT, Oey NA, Zuidervaart MM, et al. The acyl-CoA synthetase “bubblegum”(lipidosin):Further characterization and role in neuronal fatty acid β-oxidation[J]. J Biol Chem, 2003, 278(47):47070-47078.
doi: 10.1074/jbc.M310075200 URL |
| [9] |
Babbitt PC, Kenyon GL, Martin BM, et al. Ancestry of the 4-chlorobenzoate dehalogenase:analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases[J]. Biochemistry, 1992, 31(24):5594-5604.
pmid: 1351742 |
| [10] |
Iijima H, Fujino T, Minekura H, et al. Biochemical studies of two rat acyl-CoA synthetases, ACS1 and ACS2[J]. Eur J Biochem, 1996, 242(2):186-190.
pmid: 8973631 |
| [11] |
Steinberg SJ, Morgenthaler J, Heinzer AK, et al. Very long-chain acyl-CoA synthetases:human “bubblegum” represents a new family of proteins capable of activating very long-chain fatty acids[J]. J Biol Chem, 2000, 275(45):35162-35169.
doi: 10.1074/jbc.M006403200 pmid: 10954726 |
| [12] |
Shockey JM, Fulda MS, Browse JA. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism[J]. Plant Physiol, 2002, 129(4):1710-1722.
pmid: 12177484 |
| [13] |
Zhao L, Katavic V, Li F, et al. Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1(LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis[J]. Plant J, 2010, 64(6):1048-1058.
doi: 10.1111/j.1365-313X.2010.04396.x URL |
| [14] |
Lv SY, Song T, Kosma DK, et al. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1(LACS1)that has overlapping functions with LACS2 in plant wax and cutin synthesis[J]. Plant J, 2009, 59(4):553-564.
doi: 10.1111/j.1365-313X.2009.03892.x URL |
| [15] | Zhao HY, Kosma DK, Lü S. Functional role of long-chain acyl-CoA synthetases in plant development and stress responses[J]. Front Plant Sci, 2021, 12:640996. |
| [16] |
Weng H, Molina I, Shockey J, et al. Organ fusion and defective cuticle function in a Lacs1 Lacs2 double mutant of Arabidopsis[J]. Planta, 2010, 231(5):1089-1100.
doi: 10.1007/s00425-010-1110-4 pmid: 20237894 |
| [17] |
Zhao LF, Haslam TM, Sonntag A, et al. Functional overlap of long-chain acyl-CoA synthetases in Arabidopsis[J]. Plant Cell Physiol, 2019, 60(5):1041-1054.
doi: 10.1093/pcp/pcz019 URL |
| [18] |
Faergeman NJ, Black PN, Zhao XD, et al. The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular Utilization[J]. J Biol Chem, 2001, 276(40):37051-37059.
doi: 10.1074/jbc.M100884200 pmid: 11477098 |
| [19] |
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR[J]. Nucleic Acids Res, 2001, 29(9):e45.
doi: 10.1093/nar/29.9.e45 pmid: 11328886 |
| [20] | Mereshchuk A, Chew JSK, Dobson MJ. Use of yeast plasmids:transformation and inheritance assays[J]. Methods Mol Biol, 2021, 2196:1-13. |
| [21] |
Bua A, et al. Antimicrobial activity of Austroeupatorium inulaefolium(H. B. K. )against intracellular and extracellular organisms[J]. Nat Prod Res, 2018, 32(23):2869-2871.
doi: 10.1080/14786419.2017.1385014 pmid: 29017356 |
| [22] | Donadu M, Usai D, et al. In vitro activity of hybrid lavender essential oils against multidrug resistant strains of Pseudomonas aeruginosa[J]. J Infect Dev Countr, 2018, 12(1):9-14. |
| [23] |
宋燕子, 贾彬, 等. 莱茵衣藻酰基辅酶A合成酶cDNA克隆及其酵母表达[J]. 生物技术通报, 2015, 31(9):119-124.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.09.016 |
| Song YZ, Jia B, et al. cDNA cloning and yeast expression of acyl-CoA synthetase of Chlamydomonas reinhardtii[J]. Biotechnol Bull, 2015, 31(9):119-124. | |
| [24] | 李艾. 斯氏油脂酵母细胞内油脂含量的快速检测方法研究[J]. 中国油脂, 2016, 41(11):79-82. |
| Li A. Rapid determination of oil content in Lipomyces Starkeyi cells[J]. China Oils Fats, 2016, 41(11):79-82. | |
| [25] |
Wang XL, Li XB. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton[J]. Plant J, 2009, 57(3):473-486.
doi: 10.1111/j.1365-313X.2008.03700.x URL |
| [26] |
Jessen D, Roth C, Wiermer M, et al. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis[J]. Plant Physiol, 2015, 167(2):351-366.
doi: 10.1104/pp.114.250365 pmid: 25540329 |
| [27] |
Schnurr JA, Shockey JM, de Boer GJ, et al. Fatty acid export from the chloroplast. molecular characterization of a major plastidial acyl-coenzyme a synthetase from Arabidopsis[J]. Plant Physiol, 2002, 129(4):1700-1709.
pmid: 12177483 |
| [28] | Yu L, Tan X, Jiang B, et al. A peroxisomal long-chain acyl-CoA synthetase from Glycine max involved in lipid degradation[J]. PLoS One, 2014, 9(7):e100144. |
| [29] |
Aznar-Moreno JA, Venegas Calerón M, Martínez-Force E, et al. Sunflower(Helianthus annuus)long-chain acyl-coenzyme A synthetases expressed at high levels in developing seeds[J]. Physiol Plant, 2014, 150(3):363-373.
doi: 10.1111/ppl.12107 pmid: 24102504 |
| [30] |
Jessen D, Olbrich A, Knüfer J, et al. Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis[J]. Plant J, 2011, 68(4):715-726.
doi: 10.1111/j.1365-313X.2011.04722.x URL |
| [31] |
Pulsifer IP, Kluge S, Rowland O. Arabidopsis long-chain acyl-CoA synthetase 1(LACS1), LACS2, and LACS3 facilitate fatty acid uptake in yeast[J]. Plant Physiol Biochem, 2012, 51:31-39.
doi: 10.1016/j.plaphy.2011.10.003 URL |
| [32] |
McFarlane HE, Watanabe Y, et al. Golgi- and trans-Golgi network-mediated vesicle trafficking is required for wax secretion from epidermal cells[J]. Plant Physiol, 2014, 164(3):1250-1260.
doi: 10.1104/pp.113.234583 pmid: 24468625 |
| [33] |
Zhang CL, Mao K, Zhou LJ, et al. Genome-wide identification and characterization of apple long-chain Acyl-CoA synthetases and expression analysis under different stresses[J]. Plant Physiol Biochem, 2018, 132:320-332.
doi: 10.1016/j.plaphy.2018.09.004 URL |
| [34] | Zhang CL, et al. An apple long-chain acyl-CoA synthetase, MdLACS4, induces early flowering and enhances abiotic stress resistance in Arabidopsis[J]. Plant Sci, 2020, 297:110529. |
| [35] |
Cominelli E, Sala TA, Calvi D, et al. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability[J]. Plant J, 2008, 53(1):53-64.
pmid: 17971045 |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 宋志忠, 徐维华, 肖慧琳, 唐美玲, 陈景辉, 管雪强, 刘万好. 酿酒葡萄铁调节转运蛋白基因VvIRT1的克隆、表达与功能[J]. 生物技术通报, 2023, 39(8): 234-240. |
| [3] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [4] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [5] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [6] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [7] | 镐青青, 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶. 马尾松NAC转录因子基因PmNAC8的克隆及表达分析[J]. 生物技术通报, 2022, 38(4): 202-216. |
| [8] | 赵婷婷, 王俊刚, 王文治, 冯翠莲, 冯小艳, 张树珍. 甘蔗单糖转运蛋白基因ShSTP7序列分析及组织表达特征测定[J]. 生物技术通报, 2022, 38(4): 72-78. |
| [9] | 张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2022, 38(12): 184-193. |
| [10] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| [11] | 曹映辉, 胡美娟, 童妍, 张燕萍, 赵凯, 彭东辉, 周育真. 建兰ABC基因家族鉴定及其在花发育过程中的表达模式分析[J]. 生物技术通报, 2022, 38(11): 162-174. |
| [12] | 骆鹰, 谭智, 王帆, 刘晓霞, 罗小芳, 何福林. 银杏GbR2R3-MYB1基因的克隆及非生物胁迫应答分析[J]. 生物技术通报, 2022, 38(10): 184-194. |
| [13] | 孙瑞芬, 张艳芳, 牛素清, 郭树春, 李素萍, 于海峰, 聂惠, 牟英男. 向日葵HaACO1基因的表达分析及功能验证[J]. 生物技术通报, 2021, 37(9): 114-124. |
| [14] | 范亚朋, 芮存, 张悦新, 陈修贵, 陆许可, 王帅, 张红, 徐楠, 王晶, 陈超, 叶武威. 陆地棉耐碱基因GHZAT12的克隆、表达及生物信息学分析[J]. 生物技术通报, 2021, 37(8): 121-130. |
| [15] | 山草梅, 叶蕾, 张连虎, 况卫刚, 孙晓棠, 马建, 崔汝强. 水稻抗潜根线虫基因OsRAI1的克隆及功能分析[J]. 生物技术通报, 2021, 37(7): 146-155. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||